Protein Chemistry and Amino Acids
Protein Chemistry : Overworkers
PROTEINS
Proteins are undoubtedly the most versatile and functionally diverse macromolecules in living systems. They are massive, complex organic compounds that are absolutely essential for every living cell, performing the vast majority of biological tasks. Indeed, if you can imagine a job that needs doing in a cell, chances are a protein is doing it.
Think of them as the true "workhorses" of the cell. While carbohydrates are primarily for immediate energy and structural components, and lipids for membranes and long-term energy storage, proteins execute an astonishing array of functions, making life possible and dynamic.
Origin of the Term "Protein"
The word "protein" is derived from the Greek word "proteios."
- "Proteios" means "holding the first place" or "primary."
This etymology beautifully underscores their profound significance: proteins are indeed of utmost importance to life, playing a primary and central role in virtually every biological process, from molecular interactions to macroscopic tissue function.
What are Proteins?
- Most Abundant Organic Molecules: Proteins are the most abundant and functionally diverse organic macromolecules found in living systems. They make up a significant portion of a cell's dry weight (often 50-70%), underscoring their ubiquitous presence and essential roles.
- Large Molecules (Biopolymers/Macromolecules): Proteins are large, complex molecules, often referred to as biopolymers or macromolecules due to their considerable size and intricate three-dimensional structures. Their precise folding is critical for their function.
- Made of Amino Acids: The Monomeric Units: They are constructed from smaller, repeating building blocks called amino acids. There are 20 common, genetically encoded amino acids that serve as the fundamental units for protein synthesis.
- Polymers of Amino Acids: The Polypeptide Chain: Proteins are fundamentally polymers of amino acids, linked together in long, unbranched chains. This linear sequence of amino acids is called a polypeptide chain. The sequence dictates the protein's unique 3D structure and, consequently, its specific function.
- Ubiquitous Presence: Proteins are found in every part of a cell and throughout the body – in cytoplasm, organelles, membranes, extracellular matrix, fluids (e.g., blood plasma, lymph), secretions, and even excretions. In human plasma alone, over 300 different types of proteins have been identified, each with distinct roles!
- Basis of Body Structure & Function: They form the fundamental basis of body structure, from the cytoskeleton of individual cells to the collagen in our bones and skin. Moreover, they are intimately involved in most of the body's functions and life processes, orchestrating the complex machinery of life.
- DNA Dictates Sequence (Central Dogma of Molecular Biology): The specific sequence of amino acids in a protein is precisely determined by the genetic information encoded in our DNA (Deoxyribonucleic Acid). This process, known as gene expression, involves transcription of DNA into messenger RNA (mRNA) and then translation of mRNA into a polypeptide chain on ribosomes. This precise control ensures that each protein has the correct sequence for proper folding and function.
Elemental Composition: What are Proteins Made Of?
While carbohydrates and lipids primarily consist of carbon, hydrogen, and oxygen, proteins possess a broader and more distinctive elemental signature:
- Carbon (C): 50 – 55%
- Hydrogen (H): 6 – 7.3%
- Oxygen (O): 19 – 24%
- Nitrogen (N): 13 – 19% (average is approximately 16%). This consistent presence of nitrogen in all proteins is the key differentiator that sets them apart from carbohydrates and lipids. This nitrogen is primarily found in the amino groups of their amino acid building blocks.
- Sulfur (S): 0 – 4% (present in the side chains of specific amino acids like Cysteine and Methionine, which are crucial for forming disulfide bonds and maintaining protein structure).
- Phosphorus (P): While not a primary constituent of the polypeptide backbone, phosphorus can be covalently attached to proteins through post-translational modifications (e.g., phosphorylation of Serine, Threonine, or Tyrosine residues), which is a critical regulatory mechanism for protein activity. Some proteins also contain metal ions (e.g., Iron in hemoglobin, Zinc in many enzymes) as cofactors.
Functions of Proteins:
Proteins are truly the "workhorses" that carry out the cellular instructions and enable all aspects of life. Their functions are incredibly diverse and sophisticated:
Structural Support
Proteins provide the framework and strength for cells and tissues. Examples include Collagen (in skin, bone), Elastin (in blood vessels), Keratin (in hair, nails), and Actin/Tubulin (in the cytoskeleton).
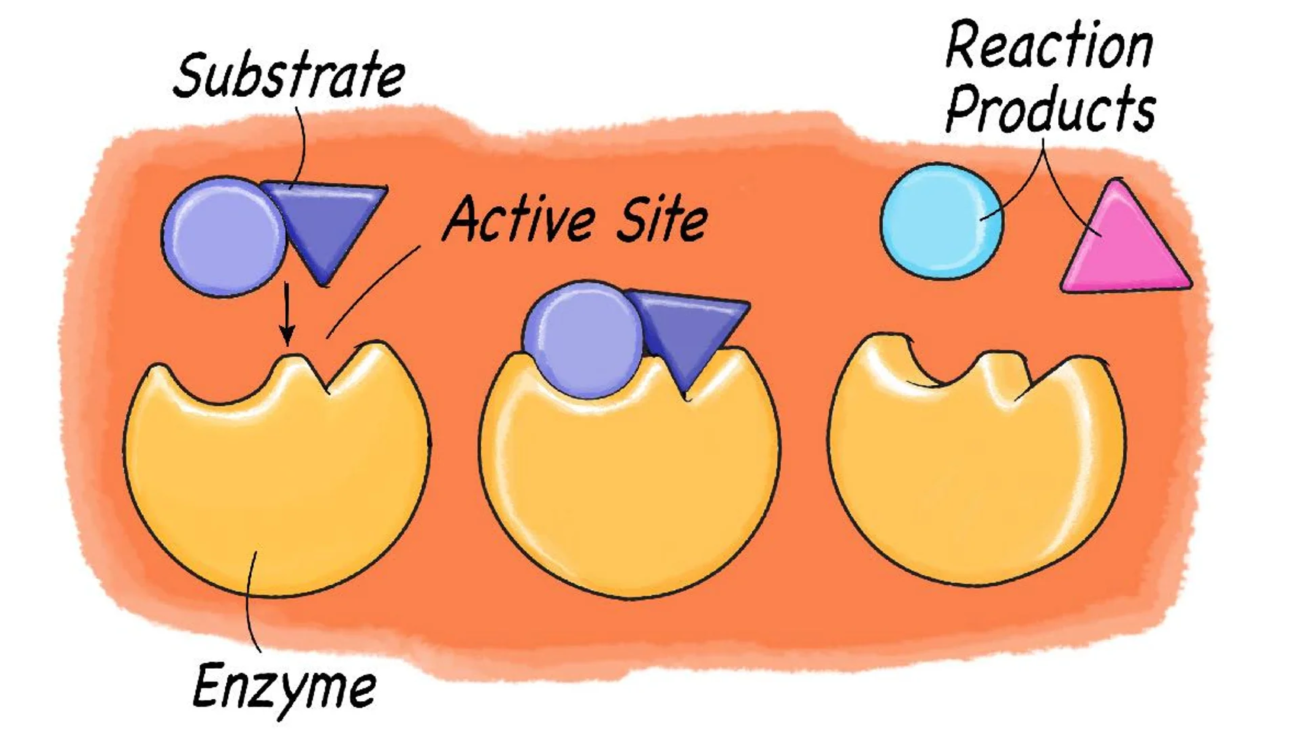
Catalysis (Enzymes)
As enzymes, proteins speed up nearly all biochemical reactions. Examples include Amylase (digests starch) and DNA Polymerase (synthesizes DNA). Deficiencies can cause metabolic diseases.
Transport and Storage
Proteins move essential molecules. Hemoglobin transports oxygen, Albumin transports fatty acids and drugs, Lipoproteins transport fats, and Transferrin transports iron. Ferritin stores iron inside cells.
Movement
Contractile proteins enable all forms of biological movement. Actin and Myosin power muscle contraction, while Dynein and Kinesin move cargo within cells and power cilia and flagella.
Regulation & Signaling
Proteins regulate physiological processes. Examples include protein hormones like Insulin, cell surface Receptors that transmit signals, and Transcription Factors that control gene expression.
Immune Defense
Proteins protect the body from pathogens. Antibodies (Immunoglobulins) recognize and neutralize foreign invaders, while Cytokines and Complement proteins coordinate the immune response.
Fluid Balance & Clotting
Plasma proteins like Albumin maintain osmotic pressure, preventing tissue edema. Coagulation factors like Fibrinogen and Thrombin are essential for blood clotting and preventing blood loss after injury.
Energy Source
While not their primary function, proteins can be broken down into amino acids and used for energy during times of starvation or when other energy stores are depleted, through processes like gluconeogenesis.
Anatomy of Amino Acids: The Building Blocks of Proteins
Remember the functional group Amino? Indeed, it's central to these vital molecules!
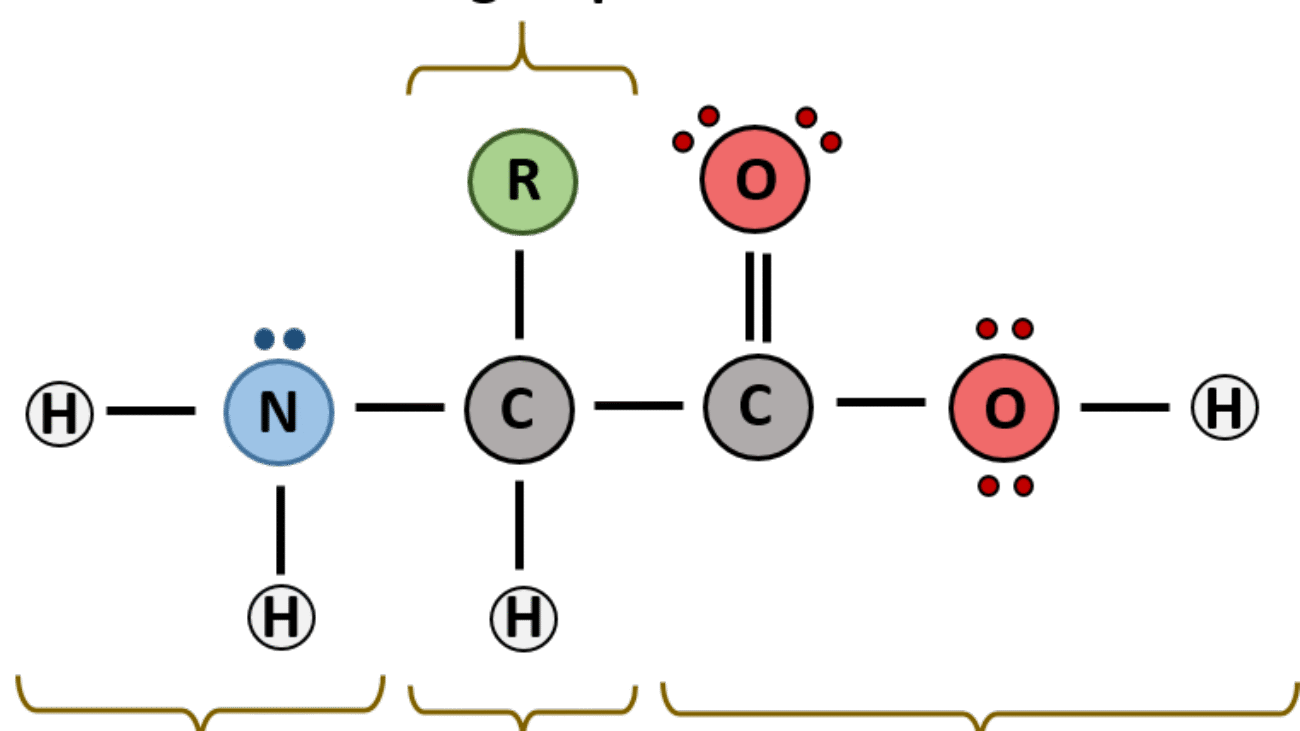
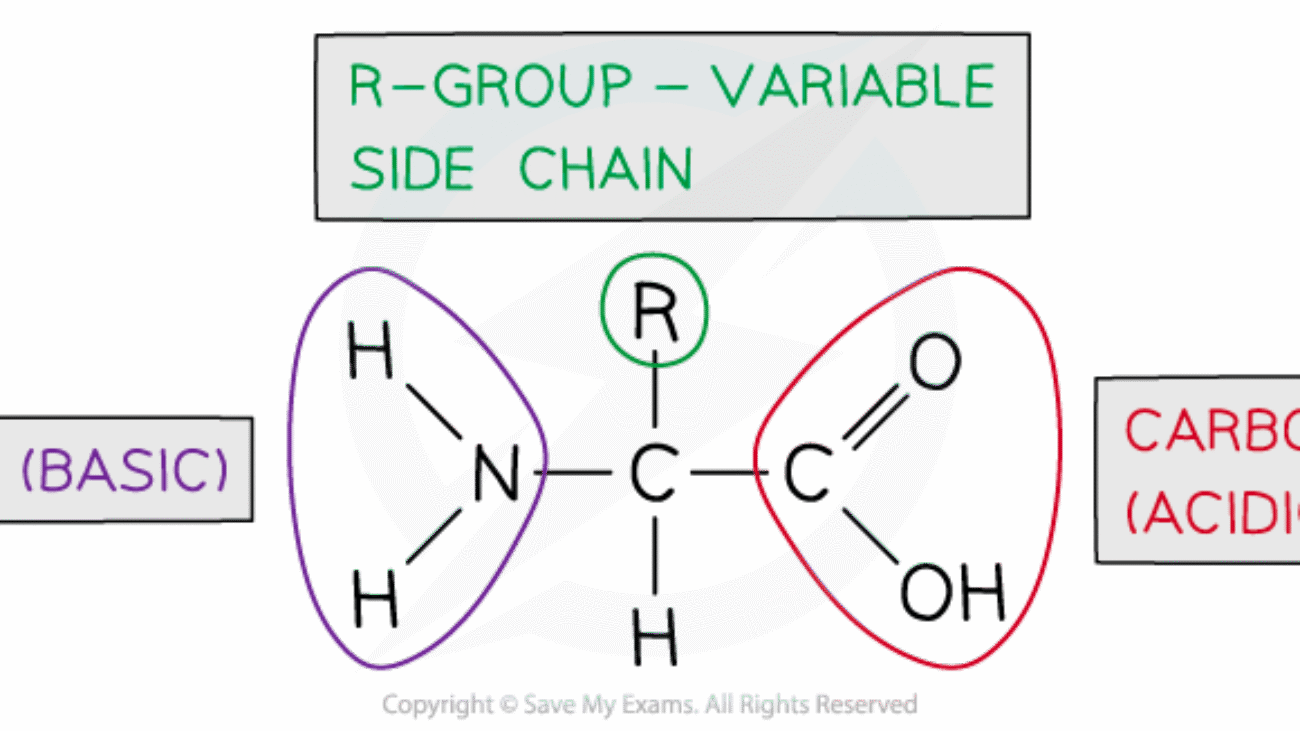
An amino acid is an organic molecule characterized by its unique chemical structure: it features a central carbon atom (the α-carbon) covalently bonded to four distinct groups:
- A basic amino group (−NH2)
- An acidic carboxyl group (−COOH)
- A hydrogen atom (−H)
- An organic R group (or side chain) that is unique to each specific amino acid.
The term amino acid is short for α-amino carboxylic acid, emphasizing the attachment of both the amino and carboxyl groups to the same carbon atom (the α-carbon).
The Basic Shape of Every Amino Acid (The "Amino Acid Blueprint")
Every single one of the 20 common genetically encoded amino acids shares a very similar basic blueprint:
- A central carbon (C) atom, called the alpha (α)-carbon. This carbon is the structural heart of the amino acid.
- Attached to this central α-carbon are four different chemical groups:
- An "Amino Group" (−NH2): This group contains nitrogen and is characterized by its basic properties. At physiological pH (the normal pH inside the body, approximately 7.4), the amino group is protonated, carrying a positive electrical charge (−NH3+).
- A "Carboxyl Group" (−COOH): This group contains carbon and oxygen and is characterized by its acidic properties. At physiological pH, the carboxyl group is deprotonated, carrying a negative electrical charge (−COO−).
- A "Hydrogen Atom" (−H): A single hydrogen atom that completes the valency of the α-carbon.
- A "Side Chain" (or "R-Group"): This is the most critical and defining part that makes each amino acid unique. The R-group can range from a single hydrogen atom (as in glycine) to complex cyclic structures. It is the R-group's specific chemical properties (e.g., size, shape, charge, polarity, hydrogen-bonding capacity) that determine the overall chemical behavior of the amino acid and, ultimately, the protein it forms.
At the normal pH inside the body (physiological pH, ~7.4), the amino group typically carries a positive charge (NH3+), and the carboxyl group carries a negative charge (COO−). This means that a single amino acid, even with both positive and negative parts, can have an overall neutral charge. When a molecule possesses both a positive and a negative charge, it's called a zwitterion.
- Zwitterion: A neutral molecule that has both a positive and a negative charge within its structure. Amino acids exist predominantly as zwitterions at physiological pH. It's important to distinguish this from a molecule that has no charge at all; a zwitterion has charges, but they balance each other out for an overall neutral molecule.
- Physiological pH (7.4): At this pH, the acidic carboxyl group is dissociated, forming a negatively charged carboxylate ion (COO−). Simultaneously, the basic amino group is protonated, forming a positively charged ammonium ion (NH3+). This simultaneous presence of opposite charges within the same molecule defines the zwitterionic state.
Monomers and Polymers: From Amino Acids to Proteins
To build the long, complex chains of proteins, we need individual building blocks. These single parts are called monomers. In this case, amino acids are the fundamental monomers. When many amino acid monomers come together and link chemically, they form polymers, which are the proteins.
In simple terms: Amino acids are the building blocks, and proteins are the intricate structures built from these blocks.
How Amino Acids Connect: The Peptide Bond
Amino acids are the fundamental monomer units that link together to form polypeptides, which then fold into functional proteins. There are 20 common amino acids that are genetically encoded and found in most proteins, although many other non-proteinogenic amino acids exist in nature (e.g., modified amino acids, neurotransmitters like GABA).
- Peptide Bond: Amino acids link together by a special type of covalent chemical bond called a peptide bond. This bond is the backbone of all proteins.
- Peptides: When two or more amino acids are linked together by peptide bonds, they form a molecule called a peptide. Peptides are essentially short chains of amino acids.
- If two amino acids combine, it's a dipeptide.
- If three amino acids combine, it's a tripeptide.
- If four amino acids combine, it's a tetrapeptide.
- If five amino acids combine, it's a pentapeptide.
- For chains with a relatively small number of amino acids (typically 2 to about 20-30), they are generally referred to as oligopeptides (from the Greek "oligo," meaning "few"). Examples include hormones like oxytocin or vasopressin, and some toxins.
- For longer chains of many amino acids (typically more than 30-50, extending to hundreds or thousands), they are called polypeptides (from the Greek "poly," meaning "many").
- From Polypeptide to Protein: A protein is a functional biological macromolecule made up of one or more polypeptide chains that have folded into a very specific, unique, and stable three-dimensional shape. This precise 3D structure is absolutely critical for its biological activity. So, a polypeptide is the linear chain of amino acids, and a protein is the functional, folded molecule that might contain one or more of these chains, often stabilized by additional interactions.
Formation of a Peptide Bond: Dehydration Synthesis
A peptide bond is formed through a condensation reaction (also known as dehydration synthesis).
- During this reaction, the carboxyl group (−COOH) of one amino acid reacts with the amino group (−NH2) of another amino acid.
- A molecule of water (H2O) is removed (lost) during the process. Specifically, the hydroxyl group (−OH) from the carboxyl end and a hydrogen atom (−H) from the amino end are removed.
- This forms a strong covalent amide linkage between the carbon of the carboxyl group of the first amino acid and the nitrogen of the amino group of the second amino acid. This new C-N bond is the peptide bond.
- Structure of the Peptide Bond: The peptide bond exhibits partial double-bond character due to resonance, which makes it rigid and planar. This rigidity is important for the structural integrity of the polypeptide backbone, limiting rotation around the bond and influencing the overall protein folding.
Breaking a Peptide Bond: Hydrolysis
A peptide bond can be broken through a reaction called hydrolysis.
- Hydrolysis involves the addition of a water molecule (H2O), which then breaks the covalent peptide bond. This process essentially reverses dehydration synthesis, regenerating the free carboxyl group and free amino group.
- In biological systems, this reaction is typically catalyzed by specific enzymes called proteases (or peptidases), which are essential for protein digestion, turnover, and regulation.
Amino Acid Residues: The Components of a Polypeptide
When amino acids link together to form a peptide, they lose some atoms (the elements of water) in the process. The amino acid that has been incorporated into the chain, now missing those elements, is no longer a "free amino acid" with its full amino and carboxyl groups. Instead, it's now a "leftover part" or a "component" of the larger chain. For this reason:
- An amino acid unit in a peptide (or protein) is often called a "residue."
- Proteins are polymers of amino acid residues, with each residue joined to its neighbor by a specific type of covalent bond called a peptide bond.
Properties of Amino Acids
- Solubility: Most amino acids are soluble in water and insoluble in non-polar organic solvents. This is primarily due to their charged (zwitterionic) nature and the presence of polar functional groups within their R-chains, allowing them to form strong hydrogen bonds with water molecules.
- Melting Points: They melt at higher temperatures (typically > 200°C) compared to other organic compounds of similar size. This high melting point is a direct consequence of their zwitterionic nature, where strong electrostatic forces (ionic bonds) exist between the oppositely charged groups of adjacent amino acid molecules in their crystalline state, requiring significant energy to break.
- Taste:
- Sweet: Glycine, Alanine, Valine (and some other small, non-polar or uncharged polar amino acids). This is due to their ability to bind to taste receptors.
- Bitter: Arginine, Isoleucine, Phenylalanine (often larger, more hydrophobic, or basic amino acids).
- Tasteless: Leucine (and some others).
- Umami: Glutamate (monosodium glutamate, MSG, is a common flavor enhancer).
- Note: The taste profiles are complex and depend on interactions with specific taste receptors.
- Optical Isomers (Stereoisomerism):
- All amino acids, except glycine, possess an asymmetric (chiral) α-carbon atom. A chiral carbon is bonded to four different groups.
- This chirality gives rise to optical isomers (enantiomers), which are non-superimposable mirror images of each other. These are designated as D- and L-stereoisomers.
- Nearly all biological compounds with a chiral center occur naturally in only one stereoisomeric form.
- The amino acid residues in proteins are exclusively L-stereoisomers. While D-amino acids exist in nature (e.g., in bacterial cell walls, some peptide antibiotics), they are generally not incorporated into proteins during ribosomal synthesis in higher organisms. This strict stereospecificity is fundamental to protein structure and function.
- Glycine is the exception because its R-group is simply a hydrogen atom, making its α-carbon bonded to two identical hydrogen atoms, hence it is achiral.
- Ampholytes or Zwitterions (Amphoteric Nature):
- Amino acids are ampholytes, meaning they contain both an acidic group (the carboxyl group, −COOH) which can donate a proton, and a basic group (the amino group, −NH2) which can accept a proton. This allows them to act as both an acid and a base depending on the pH of the surrounding medium.
- As discussed, at physiological pH, they exist as zwitterions, bearing both a positive (NH3+) and a negative (COO−) charge, resulting in an overall neutral molecule.
- The isoelectric point (pI) is the specific pH at which an amino acid (or protein) exists predominantly as a zwitterion, with an equal number of positive and negative charges, resulting in a net charge of zero. At its pI, an amino acid will not migrate in an electric field.
- Amphoteric Nature: This ability to act as both an acid and a base is crucial for proteins to function as buffers in biological systems, helping to resist changes in pH and maintain the narrow pH range required for cellular processes.
Ionized Nature of Amino Acid (Diagrammatic Representation)
R
|
H₂N − C − COOH (General form - often drawn this way for simplicity,
| but not how it primarily exists in solution)
H
At highly acidic pH (low pH, excess H+):
- The carboxyl group is protonated (−COOH).
- The amino group is protonated (−NH3+).
- Overall charge: Cationic (net positive charge).
R
|
+H₃N − CH − COOH (Cationic form at low pH)
At physiological pH (neutral pH, ~7.4):
- The carboxyl group is deprotonated (−COO−).
- The amino group is protonated (−NH3+).
- Overall charge: Zwitterionic (net neutral charge).
R
|
+H₃N − CH − COO− (Zwitterion or dipolar ion at physiological pH)
At highly basic pH (high pH, low H+):
- The carboxyl group is deprotonated (−COO−).
- The amino group is deprotonated (−NH2).
- Overall charge: Anionic (net negative charge).
R
|
H₂N − CH − COO− (Anionic form at high pH)
Classification of the 20 Common Amino Acids (Based on R-Groups)
As we discussed, the "Side Chain" or "R-Group" is the only part that varies among the 20 common amino acids found in proteins. These R-groups have different chemical properties that dictate the amino acid's behavior and, consequently, the protein's overall structure and function.
We can classify these 20 amino acids into several groups based on the polarity and charge of their R-groups at physiological pH (around 7.4).
Group 1: Amino Acids with Nonpolar, Aliphatic R-Groups
These R-groups are generally "water-fearing" (hydrophobic) because they consist mainly of hydrocarbons (carbon and hydrogen atoms), which do not readily form hydrogen bonds with water. They tend to cluster together in the interior of proteins, away from the aqueous environment.
| Name | 3-Letter | 1-Letter | Structure of R-Group | Key Characteristics |
|---|---|---|---|---|
| Glycine | Gly | G | -H (just a hydrogen atom) | Smallest & simplest. Only non-chiral amino acid. Allows for great flexibility in protein structure due to its small size. |
| Alanine | Ala | A | -CH₃ (methyl group) | Small, unreactive. Contributes to the hydrophobic core of proteins. |
| Valine | Val | V | -CH(CH₃)₂ (isopropyl group) | Branched hydrocarbon chain. More hydrophobic than Alanine. |
| Leucine | Leu | L | -CH₂CH(CH₃)₂ (isobutyl group) | Branched hydrocarbon chain. Very hydrophobic. Common in the interior of proteins. |
| Isoleucine | Ile | I | -CH(CH₃)CH₂CH₃ (sec-butyl group) | Branched hydrocarbon chain. Stereoisomer of Leucine (same atoms, different arrangement). Very hydrophobic. |
| Methionine | Met | M | -CH₂CH₂SCH₃ (contains a sulfur atom) | Contains a sulfur atom (thioether linkage), but it's largely nonpolar. Always the first amino acid in a newly synthesized polypeptide chain (start codon). |
| Proline | Pro | P | -CH₂CH₂CH₂- (cyclic structure) | Unique cyclic structure where its R-group is bonded to both the α-carbon and the α-amino group, forming a rigid ring. Causes "kinks" in polypeptide chains. Often found in turns. |
Group 2: Amino Acids with Aromatic R-Groups
These R-groups contain bulky ring structures, which makes them generally hydrophobic. They can also absorb UV light at 280 nm, a property used to quantify proteins.
| Name | 3-Letter | 1-Letter | Structure of R-Group | Key Characteristics |
|---|---|---|---|---|
| Phenylalanine | Phe | F | -CH₂- (phenyl group) | Very hydrophobic due to the bulky phenyl ring. |
| Tyrosine | Tyr | Y | -CH₂- (phenyl group with -OH) | Aromatic ring with a hydroxyl (-OH) group. The -OH group can form hydrogen bonds, making it slightly more polar than Phenylalanine. Can be phosphorylated, important for cell signaling. |
| Tryptophan | Trp | W | -CH₂- (indole group, double ring with N) | Largest and most hydrophobic aromatic amino acid. Indole ring can form hydrogen bonds through its N-H group. Precursor to serotonin and niacin. |
Group 3: Amino Acids with Uncharged, Polar R-Groups
These R-groups contain functional groups that can form hydrogen bonds with water (like -OH, -SH, -CONH₂), making them "water-loving" (hydrophilic). They tend to be found on the surface of proteins, interacting with the aqueous environment.
| Name | 3-Letter | 1-Letter | Structure of R-Group | Key Characteristics |
|---|---|---|---|---|
| Serine | Ser | S | -CH₂OH (hydroxyl group) | Contains a hydroxyl group. Can form hydrogen bonds. Can be phosphorylated, important for cell signaling. |
| Threonine | Thr | T | -CH(OH)CH₃ (hydroxyl group) | Contains a hydroxyl group. Can form hydrogen bonds. Can be phosphorylated. |
| Cysteine | Cys | C | -CH₂SH (sulfhydryl group) | Contains a sulfhydryl (-SH) group. Crucially, two Cysteine residues can form a disulfide bond (-S-S-), a strong covalent bond that stabilizes protein structure. |
| Asparagine | Asn | N | -CH₂CONH₂ (amide group) | Contains an amide group. Can form hydrogen bonds. |
| Glutamine | Gln | Q | -CH₂CH₂CONH₂ (amide group) | Contains an amide group. Can form hydrogen bonds. Longer side chain than Asparagine. |
Group 4: Amino Acids with Positively Charged R-Groups (Basic)
These R-groups contain an extra amino group or other nitrogen-containing groups that can accept a proton (H⁺) at physiological pH, making them positively charged (basic). They are very hydrophilic and are usually found on the surface of proteins.
| Name | 3-Letter | 1-Letter | Structure of R-Group | Key Characteristics |
|---|---|---|---|---|
| Lysine | Lys | K | -CH₂CH₂CH₂CH₂NH₃⁺ (primary amine) | Long hydrocarbon chain with a terminal primary amino group. Strongly basic and positively charged at neutral pH. |
| Arginine | Arg | R | -CH₂CH₂CH₂NHC(=NH)NH₂⁺ (guanidinium group) | Contains a guanidinium group, which is the most strongly basic functional group in amino acids. Always positively charged at neutral pH. |
| Histidine | His | H | -CH₂- (imidazole group) | Contains an imidazole ring. Unique in that its side chain can be either uncharged or positively charged at physiological pH (pKa near 6.0). This makes it important in enzyme active sites, where it can act as both a proton donor and acceptor. |
Group 5: Amino Acids with Negatively Charged R-Groups (Acidic)
These R-groups contain an extra carboxyl group that can donate a proton (H⁺) at physiological pH, making them negatively charged (acidic). They are very hydrophilic and are usually found on the surface of proteins, often involved in ionic interactions.
| Name | 3-Letter | 1-Letter | Structure of R-Group | Key Characteristics |
|---|---|---|---|---|
| Aspartate | Asp | D | -CH₂COO⁻ (carboxylic acid group) | Contains a second carboxyl group. Negatively charged at neutral pH. Often participates in ionic bonds and salt bridges. |
| Glutamate | Glu | E | -CH₂CH₂COO⁻ (carboxylic acid group) | Contains a second carboxyl group. Negatively charged at neutral pH. Longer side chain than Aspartate. |
Understanding and Naming Peptide Sequences
A peptide or protein sequence is the specific linear order of amino acids linked together by peptide bonds. There are very specific conventions for how these sequences are written and read, which are essential for clear, unambiguous communication in biochemistry and molecular biology.
The Directionality of Peptides: N-terminus and C-terminus
Every peptide or polypeptide chain exhibits a distinct directionality, meaning it has a defined "start" and an "end." This intrinsic polarity is fundamental to how proteins are synthesized, fold, and function.
- Amino-terminal end (N-terminus): This is conventionally considered the beginning of the peptide chain. It is characterized by having a free, unbonded α-amino group (−NH3+) at one end of the first amino acid in the sequence. By convention, this end is always written on the left side of the sequence.
- Carboxyl-terminal end (C-terminus): This is conventionally considered the end of the peptide chain. It is characterized by having a free, unbonded α-carboxyl group (−COO−) at the other end of the last amino acid in the sequence. By convention, this end is always written on the right side of the sequence.
Reading Peptide Sequences
Peptide sequences are always read from left to right, starting from the N-terminus and proceeding sequentially towards the C-terminus.
Each amino acid unit within the peptide chain, after forming peptide bonds, is referred to as an amino acid residue. This term emphasizes that each amino acid has lost the elements of water (a hydrogen atom from its amino group and a hydroxyl group from its carboxyl group) when participating in the formation of a peptide bond. Within the chain, only the R-group and the α-carbon, along with parts of the backbone, remain.
Representing Amino Acids in a Sequence: Abbreviations
To simplify the writing and reading of often very long protein sequences, standard abbreviations are universally used for the 20 common genetically encoded amino acids:
- Three-Letter Code: Each amino acid has a unique three-letter abbreviation. These are often used when the structure is being discussed in more detail, for shorter peptides, or in academic texts to enhance readability. (e.g., Ala for Alanine, Gly for Glycine, Ser for Serine). The first letter is usually capitalized, followed by two lowercase letters.
- One-Letter Code: For very long sequences (like entire proteins or genomic sequences), a single-letter code for each amino acid is used to save space, facilitate database storage, and make sequence comparisons visually easier. This is extremely common in bioinformatics and molecular biology.
Quick reference for the 20 common amino acids and their abbreviations:
| Amino Acid | Three-Letter Code | One-Letter Code |
|---|---|---|
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartate | Asp | D |
| Cysteine | Cys | C |
| Glutamine | Gln | Q |
| Glutamate | Glu | E |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophan | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |
Note: For cases where the exact amide status is unknown or ambiguous:
- "B" can represent Asx (Aspartic acid or Asparagine).
- "Z" can represent Glx (Glutamic acid or Glutamine).
- "X" represents an unknown or unspecified amino acid.
How to "Name" or Write a Peptide Sequence
When asked to "name" a peptide or write its sequence, you list the amino acid residues in order from the N-terminus to the C-terminus, using their standard abbreviations.
- For shorter peptides: You can use three-letter codes separated by hyphens to clearly delineate each residue.
Example: Ala-Gly-Ser - For longer peptides or proteins: You primarily use one-letter codes, often written consecutively without separators, unless referring to specific segments or indicating post-translational modifications.
Example: AGS (for Ala-Gly-Ser)
Example: The sequence Asp-Lys-Gln-His-Cys-Arg-Phe can be written as DKQHCRF.
Example Practice: Determining a Peptide Sequence from its Structure
Let's carefully examine this peptide structure to determine its sequence.
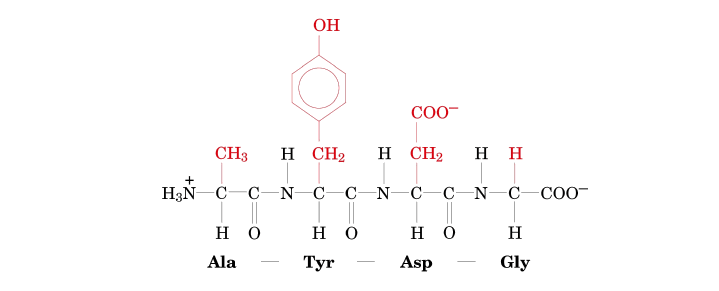
- 1st Residue (from N-terminus): R-group is −CH3.
- 2nd Residue: R-group is a phenyl group with an −OH attached to the ring (i.e., −CH2−C6H4−OH).
- 3rd Residue: R-group is −CH2CONH2.
- 4th Residue (at C-terminus): R-group is −H.
Step-by-step identification:
- Identify the N-terminus: This is on the far left, characterized by the free H3N+ group.
- Identify the C-terminus: This is on the far right, characterized by the free COO− group.
- Identify each amino acid residue from N- to C-terminus based on its R-group:
- 1st Residue (N-terminus): R-group is −CH3. This R-group corresponds to Alanine (Ala).
- 2nd Residue: R-group is a phenyl group with an −OH attached (−CH2−C6H4−OH). This R-group corresponds to Tyrosine (Tyr).
- 3rd Residue: R-group is −CH2CONH2. This R-group corresponds to Asparagine (Asn).
(−CH2CONH2 is characteristic of Asparagine.) - 4th Residue (C-terminus): R-group is −H. This R-group corresponds to Glycine (Gly).
- Write the sequence using the standard abbreviations:
- Using three-letter codes: Ala-Tyr-Asn-Gly
- Using one-letter codes: ATNG
Other Classifications of Amino Acids
Beyond the R-group classification (which is by far the most common in structural biochemistry and determines an amino acid's direct contribution to protein structure and interaction), amino acids can also be classified based on their chemical properties, nutritional requirements, and metabolic fates. These classifications provide different lenses through which to understand their roles in biology.
II. Chemical Classification
This classification often overlaps with the R-group classification (e.g., polar, nonpolar, charged) but can highlight specific chemical properties not solely related to polarity or charge. It categorizes amino acids based on the overall nature of their side chains and their behavior in solution.
- Neutral Amino Acids: These amino acids have an equal number of amino (−NH2) and carboxyl (−COOH) groups in their structure, with no additional acidic or basic groups in their R-chain that would contribute a net charge at physiological pH. Their R-groups can be either nonpolar (hydrophobic) or polar but uncharged.
Examples: Glycine, Alanine, Valine, Leucine, Isoleucine, Methionine, Proline, Phenylalanine, Tryptophan (nonpolar/hydrophobic); Serine, Threonine, Cysteine, Asparagine, Glutamine, Tyrosine (polar uncharged). - Acidic Amino Acids: These possess an additional carboxyl group (−COOH) within their R-chain, in addition to the α-carboxyl group. This extra acidic group can deprotonate at physiological pH, giving them a net negative charge at pH 7.4.
Examples: Aspartate (Asp) and Glutamate (Glu). (Note: When protonated, they are called aspartic acid and glutamic acid, respectively). - Basic Amino Acids: These contain an additional amino group (−NH2) or other nitrogenous groups capable of accepting a proton within their R-chain. These groups become protonated at physiological pH, giving them a net positive charge at pH 7.4.
Examples: Lysine (Lys), Arginine (Arg), Histidine (His). (Histidine's imidazole ring has a pKa near physiological pH, meaning it can be uncharged or positively charged depending on the exact pH). - Sulfur-Containing Amino Acids: These are characterized by the presence of sulfur atoms in their R-groups.
Examples: Cysteine (Cys) and Methionine (Met). Cysteine's thiol (−SH) group is particularly reactive and crucial for forming disulfide bonds (−S−S−), which are covalent linkages important for stabilizing protein tertiary and quaternary structures. Methionine, containing a thioether, is less reactive but often serves as the initiating amino acid in protein synthesis and as a methyl donor. - Aromatic Amino Acids: These amino acids contain an aromatic ring structure within their R-groups. These rings are generally hydrophobic and can absorb UV light, a property used in protein quantification.
Examples: Phenylalanine (Phe), Tyrosine (Tyr), Tryptophan (Trp). - Imino Acid: Technically, Proline (Pro) is often classified separately as an imino acid rather than a true amino acid. This is because its nitrogen atom (from the α-amino group) is part of a cyclic structure (a five-membered ring with the R-group), forming a secondary amine (−NH−) rather than a primary amine (−NH2). This unique structure gives proline distinct conformational properties, introducing kinks in polypeptide chains.
III. Nutritional Classification
This classification is from a dietary perspective, particularly for humans. It categorizes amino acids based on whether the human body can synthesize them de novo (from scratch) or if they must be obtained through the diet.
Essential
The body cannot synthesize these, so they must be obtained from the diet. There are 9: Histidine, Isoleucine, Leucine, Lysine, Methionine, Phenylalanine, Threonine, Tryptophan, and Valine.
Non-Essential
The body can synthesize these from other compounds, so they are not required in the diet. Examples include Alanine, Aspartate, Glycine, and Serine.
Conditionally Essential
Normally non-essential, but become essential during illness, rapid growth, or stress. Examples include Arginine, Cysteine, Tyrosine, and Glutamine.
Mnemonic (a common one, often extended): PVT TIM HALL (Phenylalanine, Valine, Threonine, Tryptophan, Isoleucine, Methionine, Histidine, Arginine, Leucine, Lysine). Note: Arginine is often considered conditionally essential, see below.
IV. Metabolic Classification
This classification categorizes amino acids based on the fate of their carbon skeletons after the amino group has been removed (a process called deamination or transamination). This dictates how the body uses them for energy production or to synthesize other crucial biomolecules.
Glucogenic
These can be converted into glucose via gluconeogenesis. Their carbon skeletons are degraded to intermediates like pyruvate or oxaloacetate. Examples include Alanine, Glycine, and Serine.
Ketogenic
These can be converted into ketone bodies or their precursors (acetyl-CoA). Leucine and Lysine are the only two amino acids that are purely ketogenic.
Both
These can be degraded into intermediates that form both glucose and ketone bodies. Examples include Isoleucine, Phenylalanine, Tyrosine, and Tryptophan.
The Biuret Test: Detecting Proteins and Peptides
The Biuret test is a classic qualitative (and semi-quantitative) chemical test used to detect the presence of proteins and peptides in a solution.
Principle:
The Biuret test specifically detects the presence of peptide bonds. It relies on the ability of copper(II) ions (Cu2+) in an alkaline solution to form a distinctive violet-colored chelate complex with compounds containing two or more peptide bonds. A single amino acid or a dipeptide will not give a positive Biuret test.
Reagents:
- Biuret reagent: This reagent typically contains:
- Dilute copper(II) sulfate (CuSO4) as the source of Cu2+ ions.
- A strong alkaline solution (e.g., sodium hydroxide, NaOH, or potassium hydroxide, KOH) to provide the necessary alkaline environment.
- Often, potassium sodium tartrate (Rochelle salt) is included to chelate the Cu2+ ions, keeping them in solution and stabilizing the complex, preventing their precipitation as copper hydroxide.
Procedure:
- Add a small amount of the sample solution (e.g., protein solution, tissue homogenate) to a clean test tube.
- Add an equal volume (or a specified ratio, typically 1:1 or 2:1 sample to reagent) of Biuret reagent to the test tube.
- Mix the contents well (gently shaking or inverting) and allow it to stand for a few minutes (e.g., 5-30 minutes) at room temperature for the color to develop.
- Observe for a color change.
Results:
- Positive Result: The solution develops a violet or purple color. This clearly indicates the presence of proteins or peptides containing at least two peptide bonds. The intensity of the violet color is generally proportional to the number of peptide bonds present, and thus, often to the concentration of protein in the sample. A pinkish-purple color may indicate shorter peptides.
- Negative Result: The solution remains blue (the original color of the copper sulfate in the reagent). This indicates the absence of significant amounts of protein or peptides containing sufficient peptide bonds to form the complex. Individual amino acids or dipeptides will yield a negative result.
Applications:
- Detecting proteins in various biological solutions (e.g., blood plasma, cell lysates, culture media).
- Estimating protein concentration (when performed quantitatively using a spectrophotometer to measure absorbance at 540 nm, by comparing to a standard curve).
- Monitoring protein purification steps, to track the presence and enrichment of protein.
- Clinical diagnostics (e.g., historically used to detect protein in urine, though more specific and sensitive tests are typically employed today).
- Educational demonstrations in biochemistry and biology laboratories.
The Four Levels of Protein Structure
Proteins are not just linear chains of amino acids; they fold into precise, intricate three-dimensional structures that are absolutely essential for their biological function. This complex folding process can be described at four hierarchical levels.
1. Primary Structure (1° Structure)
- Definition: The primary structure is the simplest and most fundamental level, referring to the exact linear sequence of amino acids in a polypeptide chain. It's akin to the order of letters in a word or sentence.
- Bonding: The only covalent bonds involved in defining the primary structure are the peptide bonds that link adjacent amino acid residues. These are strong amide bonds formed between the carboxyl group of one amino acid and the amino group of the next, with the elimination of water.
- Significance: The primary structure is the most crucial level of protein structure because it dictates all subsequent levels of folding. The unique sequence of amino acids contains all the intrinsic information necessary for the polypeptide chain to spontaneously (or with the aid of chaperones) fold into its stable, functional three-dimensional form.
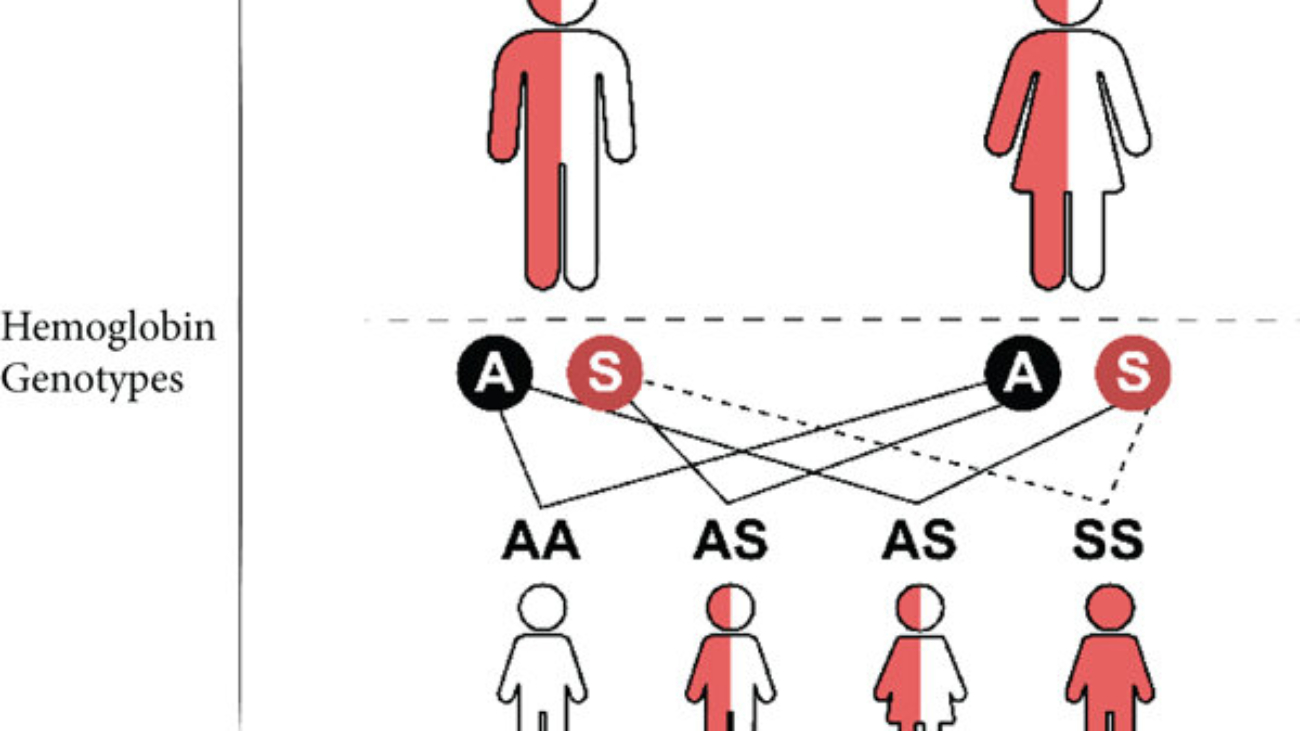
- A change in even a single amino acid (due to a gene mutation) can drastically alter the protein's higher-order structure and, consequently, its function. A classic example is sickle cell anemia, where a single amino acid substitution (Glutamate to Valine) in the beta-globin chain of hemoglobin leads to profound changes in red blood cell shape and oxygen transport capacity.
- Information Content: This level essentially contains the genetic "blueprint" or all the information needed for the protein to fold correctly into its native (functional) state.
2. Secondary Structure (2° Structure)
- Definition: Secondary structure refers to localized, regularly repeating conformations of the polypeptide chain. These structures are formed by hydrogen bonding solely between the atoms of the polypeptide backbone (specifically, the carbonyl oxygen and the amide hydrogen), not involving the R-groups at this level. These hydrogen bonds form regularly between residues that are close in the primary sequence.
- Stabilization: These structures are stabilized by an extensive network of hydrogen bonds between the carbonyl oxygen (C=O) of one peptide bond and the amide hydrogen (N-H) of another peptide bond.
Key Types: The two most common, stable, and well-defined types of secondary structure are:
a. Alpha-Helix (α-helix):
- Shape: A coiled, spiral structure resembling a right-handed screw (helical turn).
- Stabilization: Formed by hydrogen bonds that occur regularly between the carbonyl oxygen of residue n and the amide hydrogen of residue n+4 (i.e., four amino acids away along the backbone). These hydrogen bonds run roughly parallel to the helix axis.
- Characteristics:
- The R-groups of the amino acids project outward from the helix, minimizing steric hindrance and influencing interactions with the surrounding environment or other parts of the protein.
- Common in both globular and fibrous proteins.
- Certain amino acids like Proline (due to its rigid ring structure and lack of an available amide hydrogen for hydrogen bonding within the helix) and amino acids with bulky or similarly charged R-groups can disrupt alpha-helices.
- Examples: Abundant in keratin (in hair, nails, wool), and many globular proteins like myoglobin (an oxygen-storage protein).
b. Beta-Sheet (β-sheet):
- Shape: A pleated, sheet-like structure (hence "pleated sheet") formed by two or more extended polypeptide segments, called beta-strands, running alongside each other.
- Stabilization: Formed by hydrogen bonds between the carbonyl oxygen of one beta-strand and the amide hydrogen of an adjacent beta-strand. These hydrogen bonds run roughly perpendicular to the direction of the polypeptide chains.
- Orientation: Beta-sheets can be configured in two main ways:
- Parallel: Adjacent beta-strands run in the same N-to-C direction.
- Antiparallel: Adjacent beta-strands run in opposite N-to-C directions. Antiparallel sheets are generally considered more stable due to more optimal linear hydrogen bond geometry.
- Characteristics: The R-groups extend alternately above and below the plane of the sheet.
- Examples: Found in silk fibroin (gives silk its strength), and many proteins involved in immune responses or structural support like fatty acid binding proteins.
3. Tertiary Structure (3° Structure)
- Definition: Tertiary structure is the overall, elaborate three-dimensional shape or conformation of a single polypeptide chain. It describes how all the secondary structural elements (α-helices, β-sheets, and less ordered regions like turns and loops), along with random coil segments, are precisely folded and arranged relative to one another to form a compact, functional protein domain or a whole protein.
- Bonding/Interactions: Tertiary structure is stabilized by a diverse array of interactions, primarily non-covalent, occurring between the R-groups of amino acids. These interactions occur between amino acids that may be far apart in the primary sequence but are brought into close proximity by the folding process. Key interactions include:
- Hydrophobic Interactions: The primary driving force for protein folding. Nonpolar (hydrophobic) R-groups tend to cluster together in the interior of the protein, away from the aqueous cellular environment. This minimizes their contact with water and maximizes the entropy of the water molecules, leading to a more stable structure.
- Ionic Interactions (Salt Bridges): Electrostatic attractions between oppositely charged R-groups (e.g., between the negatively charged carboxyl group of an Aspartate and the positively charged amino group of a Lysine).
- Hydrogen Bonds: Formed between polar uncharged R-groups (e.g., between Serine's hydroxyl and Asparagine's amide group), or between polar R-groups and backbone atoms not already involved in secondary structure.
- Van der Waals Forces: Weak, transient attractive forces that arise from temporary fluctuations in electron distribution, occurring between all atoms that are in close proximity. While individually weak, their cumulative effect can be significant in the densely packed interior of a protein.
- Disulfide Bonds (Covalent): Unique strong covalent bonds formed by the oxidation of the sulfhydryl (−SH) groups of two Cysteine residues. These act as "molecular staples" to provide significant structural stability, particularly common in extracellular proteins exposed to oxidizing environments.
- Significance: The tertiary structure is paramount for the protein's biological function. The specific 3D arrangement creates the precise architecture necessary for:
- Active sites in enzymes for substrate binding and catalysis.
- Binding sites for ligands, cofactors, or other proteins.
- Structural motifs essential for molecular recognition and interaction.
Fibrous vs. Globular Proteins
(A classification based on overall 3D shape, largely determined by tertiary structure)
Fibrous Proteins
- Shape/Solubility: Elongated, rod-like structures; typically insoluble in water.
- Function: Primarily provide structural support, mechanical strength, and protection.
- Stabilization: Held together by strong intermolecular forces, often including numerous disulfide bonds.
- Examples: Keratin (hair, nails), Fibroin (silk), Collagen (connective tissue), Myosin (muscle).
Globular Proteins
- Shape/Solubility: Compact, spherical shape; typically soluble in water, forming colloids.
- Function: Perform diverse, dynamic roles like catalysis, transport, regulation, and immune defense.
- Stabilization: Maintained by non-covalent interactions, with hydrophobic R-groups buried inside.
- Examples: Albumin, Globulins (antibodies), Myoglobin, Insulin.
4. Quaternary Structure (4° Structure)
- Definition: Quaternary structure refers to the arrangement and interactions of multiple polypeptide chains (individual subunits) to form a larger, functional protein complex. It describes how these separate polypeptide units assemble in three-dimensional space.
- Important Note: Not all proteins have quaternary structure; it is only present in multi-subunit proteins (oligomeric proteins). Monomeric proteins (single polypeptide chain) have only primary, secondary, and tertiary structures.
- Bonding/Interactions: Similar to tertiary structure, quaternary structure is stabilized primarily by various non-covalent interactions between the R-groups of amino acids located at the interfaces of the different polypeptide chains. These include:
- Hydrophobic interactions
- Ionic interactions (salt bridges)
- Hydrogen bonds
- Van der Waals forces
- In some cases, disulfide bonds can also form between different polypeptide chains (interchain disulfide bonds), covalently linking them within the quaternary structure.
- Significance: The formation of quaternary structure often confers several advantages:
- Increased Complexity and Function: Allows for more intricate and highly regulated biological functions, often involving cooperativity or allosteric regulation.
- Enhanced Stability: Often enhances the overall stability and resistance to denaturation of the protein complex.
- Cooperation (Allostery): In some proteins (a prime example being hemoglobin), the binding of a ligand (like oxygen) to one subunit can induce conformational changes that influence the binding affinity or catalytic activity of other subunits within the same complex. This phenomenon is called allostery, crucial for finely tuning biological processes.
- Example: Hemoglobin, the oxygen-carrying protein in red blood cells, is a classic example. It consists of four polypeptide subunits (two alpha chains and two beta chains), each binding an oxygen molecule. These four subunits interact to form the functional tetrameric protein.
Protein Folding and Denaturation
The biological function of a protein is precisely linked to its precise three-dimensional structure. The journey from a linear polypeptide chain to a biologically active, folded protein is a complex and highly regulated process known as protein folding. Conversely, the loss of this critical 3D structure, leading to loss of function, is termed denaturation.
Protein Folding
Definition: Protein folding is the spontaneous (or chaperon-assisted) process by which a newly synthesized or unfolded polypeptide chain acquires its intricate, specific, and functionally active three-dimensional conformation (its native state). This precise 3D structure is determined primarily by its primary amino acid sequence.
The "Folding Problem" and Energy Landscape:
The folding of a protein from a vast number of possible conformations to a single, stable native state is often referred to as the "folding problem." This process is generally understood in terms of an energy funnel or energy landscape:
- The unfolded polypeptide exists in a high-energy, high-entropy state with many possible conformations.
- As it folds, the protein progressively moves down an energy funnel, reducing its conformational entropy and lowering its free energy.
- The bottom of the funnel represents the native, most stable, and functional 3D structure.
- Intermediate states, or "misfolded" states, can exist, which are often thermodynamically less stable or kinetically trapped.
Driving Forces and Stabilizing Interactions for Folding:
The acquisition and maintenance of the native 3D structure are driven and stabilized by a combination of weak non-covalent interactions and, occasionally, strong covalent bonds. These interactions occur between amino acid R-groups and between backbone atoms:
- Hydrophobic Effect (The Primary Driver): This is arguably the most significant driving force for protein folding in aqueous environments.
- Mechanism: Nonpolar amino acid side chains (R-groups) tend to spontaneously cluster together in the interior of the protein, effectively "hiding" away from the surrounding aqueous (water) environment. This reduces the number of ordered water molecules that would otherwise surround these nonpolar groups (minimizing the unfavorable entropy loss of water).
- Effect: The overall result is an increase in the entropy of the solvent (water) and the formation of a compact hydrophobic core within the protein, minimizing the protein's surface area exposed to water.
- Formation of Intramolecular Hydrogen Bonds:
- Mechanism: Hydrogen bonds form extensively within the protein structure.
- Backbone-Backbone H-bonds: Critical for stabilizing secondary structures (α-helices and β-sheets) between the carbonyl oxygen (C=O) of one peptide bond and the amide hydrogen (N-H) of another.
- R-group-R-group H-bonds: Between polar uncharged amino acid side chains (e.g., Serine, Threonine, Asparagine, Glutamine).
- R-group-Backbone H-bonds: Between polar side chains and backbone atoms.
- Effect: These bonds contribute significantly to the overall stability and precise geometry of both secondary and tertiary structures.
- Ionic Interactions (Salt Bridges):
- Mechanism: Electrostatic attractions between oppositely charged R-groups of acidic amino acids (e.g., Aspartate, Glutamate) and basic amino acids (e.g., Lysine, Arginine, Histidine). These interactions often involve both charge attraction and hydrogen bonding components.
- Effect: Contribute to localized stability, particularly on the surface or within specific domains, and play a role in positioning functional groups.
- Van der Waals Interactions (London Dispersion Forces):
- Mechanism: Weak, short-range attractive forces that arise from transient, fluctuating dipoles in the electron clouds of all atoms when they are in very close proximity (typically 0.3-0.6 nm).
- Effect: Individually weak, but their cumulative effect can be substantial in the densely packed interior of a protein, where many atoms are in close contact, contributing significantly to the overall stability and packing efficiency.
- Disulfide Bonds (if present):
- Mechanism: These are strong covalent bonds formed by the oxidation of the sulfhydryl (−SH) groups of two Cysteine residues. They are often formed post-translationally in the endoplasmic reticulum (for secreted or transmembrane proteins) or in the extracellular space.
- Effect: Act as robust "molecular staples" that covalently link different parts of the polypeptide chain or even different polypeptide chains (in quaternary structures), providing significant additional stability and resistance to denaturation.
Molecular Chaperones (Chaperonins)
Definition: Molecular chaperones are a diverse and essential group of proteins that assist in the proper folding of other proteins. They do not become part of the final functional protein themselves; rather, they act as "helpers" or "escorts" in the folding process. They are particularly crucial under cellular stress conditions (like heat shock) or for newly synthesized proteins, guiding them through potentially hazardous folding pathways.
Role and Mechanisms:
- Preventing Misfolding and Aggregation:
- Mechanism: Chaperones bind specifically to exposed hydrophobic regions of nascent (newly synthesized and still folding) or partially unfolded proteins. These hydrophobic patches are normally buried in the interior of correctly folded proteins. By binding to them, chaperones prevent these sticky hydrophobic regions from interacting prematurely with other hydrophobic regions of the same or different proteins, which would lead to incorrect folding or aggregation into insoluble clumps.
- Effect: Ensures that the protein has sufficient time and a protected environment to explore proper folding pathways, preventing the formation of non-functional aggregates.
- Assisting Refolding:
- Mechanism: Some chaperones (e.g., the Hsp70 family) can bind to misfolded proteins, using ATP hydrolysis to induce conformational changes that can help pull apart aggregates or allow the misfolded protein another chance to refold correctly.
- Effect: Rescues misfolded proteins, restoring their function and preventing their accumulation, which can be toxic to the cell.
- Protecting from Stress (Heat Shock Proteins - HSPs):
- Mechanism: Many chaperones are constitutively expressed but their synthesis significantly increases in response to various cellular stresses, especially elevated temperatures. They are thus often referred to as "heat shock proteins" (HSPs). Heat stress can cause proteins to partially unfold, exposing hydrophobic regions and making them prone to aggregation. HSPs rapidly upregulate to combat this.
- Effect: HSPs act as a cellular defense mechanism, protecting existing proteins from heat-induced denaturation and facilitating the refolding of stress-damaged proteins, thereby maintaining cellular proteostasis (protein homeostasis).
Major Chaperone Families: Examples include the Hsp70 family (which binds to nascent chains), Hsp90 (involved in the maturation of signaling proteins), and chaperonins like GroEL/GroES (which provide an "isolation chamber" for protein folding).
Denaturation
Definition: Denaturation is the process by which a protein loses its specific, biologically active, native three-dimensional conformation. This loss of structure typically results in a loss of biological function.
Structural Changes:
- Denaturation primarily involves the disruption of the non-covalent interactions (hydrogen bonds, hydrophobic interactions, ionic interactions, Van der Waals forces) that stabilize the secondary, tertiary, and, if present, quaternary structures.
- Crucially, denaturation typically does not break the primary structure (peptide bonds). The amino acid sequence remains intact.
Consequences:
- Loss of Biological Activity: A denatured protein becomes biologically inactive because its specific active sites, binding domains, recognition surfaces, or structural integrity are lost or significantly altered.
- Reduced Solubility and Aggregation: The exposure of normally buried hydrophobic regions often leads to reduced solubility and a strong tendency for denatured proteins to aggregate into insoluble precipitates, which can be cytotoxic.
- Increased Susceptibility to Proteolysis: Unfolded proteins are often more susceptible to degradation by proteases.
Reversibility (Renaturation):
- Denaturation can sometimes be reversible (renaturation). If the denaturing agent is removed and the conditions are returned to normal (e.g., optimal pH, temperature), the protein may spontaneously refold into its native, functional state. This phenomenon, famously demonstrated by Christian Anfinsen with ribonuclease, showed that the primary sequence contains all the information needed for folding.
- However, severe or prolonged denaturation often leads to irreversible changes. Extensive aggregation or irreversible chemical modifications can prevent proper refolding, even after the denaturing agent is removed.
Denaturing Agents (Factors that Cause Denaturation)
Various physical and chemical agents can cause denaturation by interfering with the weak forces that maintain protein structure:
a. Heat
Mechanism: Increases kinetic energy, causing vibrations that disrupt weak non-covalent interactions like hydrogen bonds and hydrophobic interactions. Effect: Causes unfolding, often irreversibly, like cooking an egg.
b. Extreme pH
Mechanism: Alters the ionization state of acidic and basic R-groups, disrupting crucial ionic bonds (salt bridges) and hydrogen bonding patterns. Effect: Causes charge repulsion and destabilizes the native conformation.
c. Organic Solvents
Mechanism: Less polar than water, these solvents (e.g., ethanol, acetone) disrupt and dissolve the internal hydrophobic core of proteins. Effect: Weakens the hydrophobic effect, leading to unfolding and precipitation.
d. Strong Detergents
Mechanism: Amphipathic molecules (e.g., SDS) bind to and disrupt hydrophobic regions, coating the protein with charge. Effect: Leads to complete unfolding into a random coil, useful in laboratory techniques.
e. Heavy Metal Ions
Mechanism: Ions like Pb²⁺ or Hg²⁺ react strongly with sulfhydryl (-SH) groups and charged R-groups. Effect: Disrupts disulfide and ionic bonds, often causing irreversible denaturation and enzyme inactivation.
f. Chaotropic Agents
Mechanism: Small molecules (e.g., urea, guanidinium chloride) disrupt the structure of water and form H-bonds with the protein. Effect: Weakens the hydrophobic effect and disrupts internal H-bonds, causing complete unfolding.
g. Mechanical Stress
Mechanism: Vigorous shaking, grinding, or shearing applies physical force that can break weak non-covalent interactions. Effect: Causes unfolding and aggregation as exposed hydrophobic regions interact, such as when whipping egg whites.
Protein Misfolding Diseases (Consequences of Folding Errors)
Despite the cellular machinery dedicated to ensuring proper protein folding, including a battery of molecular chaperones, errors can (and do) occur.
Proteins may fail to achieve their correct native state, or they may denature and subsequently refold improperly.
The accumulation of these misfolded proteins can have profound and often devastating consequences, leading to a wide array of severe diseases, prominently featuring neurodegenerative disorders. These conditions underscore the critical link between protein structure, function, and cellular health.
Mechanism of Disease: The Unifying Principles of Misfolding Pathology
While the specific proteins and affected tissues vary, a common set of pathological mechanisms underlies most protein misfolding diseases:
- Improper Folding:
- A protein either never successfully achieves its correct, lowest-energy native state during de novo synthesis. This can be due to genetic mutations that destabilize the native fold, overwhelmed chaperone systems, or unfavorable cellular environments.
- Alternatively, a correctly folded protein might denature (lose its native structure) due to stress or aging and then refold into an alternative, incorrect, and often stable conformation that lacks biological function.
- Aggregation:
- The Exposure of Sticky Surfaces: A hallmark of misfolded proteins is the exposure of normally buried hydrophobic regions or highly aggregation-prone segments. These exposed "sticky" surfaces facilitate abnormal intermolecular interactions.
- Self-Association: Misfolded proteins tend to self-associate through these exposed regions, leading to the formation of insoluble, ordered aggregates. These aggregates can range from small, soluble oligomers (which are often the most toxic species) to large, insoluble amyloid fibrils (characterized by a cross-β sheet structure) or amorphous inclusions.
- Reduced Degradation: The tightly packed, protease-resistant nature of these aggregates often renders them resistant to the cell's normal protein degradation pathways (e.g., the proteasome and lysosome/autophagy system), leading to their accumulation.
- Cellular Toxicity and Dysfunction:
- Interference with Proteostasis: The accumulation of misfolded proteins can overwhelm and impair the cell's protein quality control (proteostasis) machinery, leading to a vicious cycle where more proteins misfold and aggregate.
- Disruption of Organelle Function: Aggregates can physically interfere with the normal functioning of vital cellular organelles such as mitochondria (impairing energy production), endoplasmic reticulum (ER stress response), and lysosomes.
- Impairment of Transport: In neurons, aggregates can disrupt axonal transport, preventing essential molecules from reaching their destinations.
- Direct Toxicity: Soluble oligomers, in particular, are hypothesized to exert direct toxic effects, for example, by perforating membranes, disrupting synaptic function, or sequestering essential cellular components.
- Inflammation: Protein aggregates can also trigger inflammatory responses, further contributing to tissue damage.
- Cell Death: Ultimately, this cascade of dysfunction leads to cellular dysfunction and eventually cell death (apoptosis or necrosis), which is particularly devastating in post-mitotic cells like neurons. In the brain, this neuronal loss manifests as the clinical symptoms of neurodegenerative diseases.
Examples of Misfolding Diseases:
a. Sickle Cell Disease (SCD)
A Classic Example of a Point Mutation Leading to Aberrant Assembly
Misfolded/Mutated Protein: Hemoglobin (Hb). Specifically, a single-point mutation converts normal hemoglobin (HbA) to sickle hemoglobin (HbS) by replacing a polar glutamate with a nonpolar valine.
Mechanism: This substitution creates a "sticky" hydrophobic patch on the surface of deoxy-HbS. Under low oxygen conditions, these patches cause HbS molecules to polymerize into long, rigid, insoluble fibers that distort red blood cells into a rigid sickle shape.
Effect: The sickled cells are fragile (causing anemia) and rigid, leading to blockage of small blood vessels (vaso-occlusive crises), intense pain, and organ damage. It is a prime example of how a single amino acid change can have catastrophic physiological consequences.
b. Alzheimer's Disease (AD)
A Dual-Protein Pathology
Misfolded Proteins: Primarily involves two proteins: Beta-amyloid (Aβ) and Tau.
Mechanism: Aβ is a peptide that misfolds and aggregates extracellularly to form insoluble amyloid plaques between neurons. The Tau protein becomes hyperphosphorylated, detaches from microtubules, and aggregates intracellularly to form neurofibrillary tangles (NFTs) inside neurons.
Effect: The accumulation of both plaques and tangles is thought to cause widespread neuronal dysfunction and death, leading to progressive cognitive decline, severe memory loss, and dementia.
c. Parkinson's Disease (PD)
Synucleinopathy
Misfolded Protein: Alpha-synuclein, a protein involved in synaptic vesicle regulation.
Mechanism: Alpha-synuclein misfolds and aggregates into intracellular inclusions called Lewy bodies and Lewy neurites. These aggregates primarily affect dopaminergic neurons in the substantia nigra region of the brain.
Effect: The progressive loss of these dopamine-producing neurons leads to a severe dopamine deficiency, causing the characteristic motor symptoms of Parkinson's, including tremor, rigidity, slowness of movement (bradykinesia), and postural instability.
d. Prion Diseases (TSEs)
Transmissible Spongiform Encephalopathies
Misfolded Protein: Prion protein (PrP). This disease is unique because the misfolded protein itself is infectious.
Mechanism: A normal cellular protein (PrPC) misfolds into an abnormal, protease-resistant isoform (PrPSc). This infectious PrPSc then acts as a template, forcing other normal PrPC molecules to adopt the misfolded conformation in a self-propagating chain reaction.
Effect: The accumulation of PrPSc aggregates causes widespread neuronal death and a "spongiform" (vacuolated) appearance in the brain, leading to rapidly progressive and fatal neurodegeneration. Examples include Creutzfeldt-Jakob Disease (CJD) in humans and "Mad Cow Disease" (BSE) in cattle.
e. Cystic Fibrosis (CF)
A Quality Control Error
Misfolded Protein: Cystic Fibrosis Transmembrane Conductance Regulator (CFTR), a chloride ion channel.
Mechanism: A common mutation (ΔF508) causes the CFTR protein to misfold. While it might still be partially functional, the cell's own quality control machinery in the endoplasmic reticulum recognizes the misfolded protein and targets it for premature degradation before it can reach the cell membrane.
Effect: The lack of functional CFTR channels at the cell surface impairs chloride ion transport, leading to thick, sticky mucus in the lungs, pancreas, and other organs, causing chronic infections, respiratory failure, and malabsorption.
Clinical Case Scenario: Sickle Cell Anemia
A 2-year-old boy from Mukono district is admitted to the hospital presenting with a constellation of acute symptoms: recurrent, excruciating severe bone pain affecting his hands, feet, and sternum for the past 3 days, accompanied by noticeable jaundice and profound fatigue. His parents report previous, similar episodes.
Laboratory findings on admission reveal:
- Haemoglobin: 6.2 g/dL (significantly below the normal range of 11-16 g/dL), indicative of severe anemia.
- Peripheral blood smear: Microscopic examination strikingly shows numerous sickled red blood cells, elongated and crescent-shaped, alongside normal discocytes.
- Liver function tests: Markedly elevated bilirubin, explaining the jaundice.
- Haemoglobin electrophoresis: Confirms the presence of a significantly increased percentage of sickled haemoglobin (HbS), with a reduced percentage of normal adult haemoglobin (HbA).
Based on these findings, a diagnosis of Vaso-occlusive crisis and severe anemia due to Sickle Cell Disease was made.
(a) Explain in detail the amino acid change that occurs in this patient's haemoglobin, highlighting the nature of the amino acids involved and the chemical basis of the mutation.
Detailed Explanation of the Amino Acid Change:
The genetic basis of Sickle Cell Disease (SCD) in this patient, as confirmed by the presence of HbS, lies in a single-point mutation within the gene encoding the beta-globin chain of hemoglobin. This seemingly minor alteration in the DNA sequence triggers a profound change at the protein level:
- Genetic Mutation: The primary cause is a substitution of a single nucleotide base within the DNA. Specifically, the triplet codon GAG (which codes for Glutamate) is mutated to GTG (which codes for Valine). This alteration in the genetic code is then transcribed into mRNA, leading to a changed codon from GAG to GUG.
- Amino Acid Substitution: This altered mRNA codon (GUG) during translation directs the ribosome to incorporate Valine instead of Glutamate at the sixth position of the beta-globin polypeptide chain.
- Nature of the Amino Acids Involved:
- Glutamate (Glu, E): In its physiological ionized state, glutamate is a polar, negatively charged (acidic) amino acid. Its side chain contains a carboxyl group (−COOH) that is deprotonated to −COO− at neutral pH, making it hydrophilic and capable of forming ionic bonds (salt bridges) and hydrogen bonds. Typically, glutamate residues are found on the surface of soluble proteins, interacting favorably with the aqueous cellular environment.
- Valine (Val, V): Valine, in contrast, is a nonpolar, hydrophobic amino acid. Its side chain consists of a branched hydrocarbon chain, which does not interact favorably with water. Consequently, valine residues are typically buried in the hydrophobic core of folded proteins, away from the aqueous environment.
- Chemical Basis of the Mutation and its Impact on Protein Surface:
- The crucial chemical change is the replacement of a hydrophilic, negatively charged amino acid (Glutamate) with a hydrophobic, uncharged amino acid (Valine) at a critical, solvent-exposed position on the surface of the beta-globin protein.
- This substitution creates a newly exposed hydrophobic patch on the surface of the beta-globin subunit when hemoglobin is in its deoxy (deoxygenated) state. This hydrophobic region is normally absent in HbA, where the glutamate residue at this position would facilitate favorable interactions with water. This subtle change in surface chemistry is the initial molecular trigger for the pathogenesis of SCD.
(b) Describe how this amino acid change affects haemoglobin function at the molecular level and leads to the clinical manifestations observed.
Molecular Mechanism and Clinical Manifestations:
The single amino acid substitution of Valine for Glutamate at position 6 of the beta-globin chain profoundly alters the molecular behavior of hemoglobin S (HbS), particularly under conditions of low oxygen. This chain of events directly explains the patient's clinical presentation:
- Conformational Change upon Deoxygenation:
- Normal hemoglobin (HbA) exists as a tetramer of two alpha (α) and two beta (β) subunits. Its affinity for oxygen changes with its conformational state: the "R-state" (relaxed, oxygenated) has high affinity, while the "T-state" (tense, deoxygenated) has low affinity.
- The Valine substitution in HbS has little effect when oxygen is bound (oxy-HbS). However, upon deoxygenation (e.g., when red blood cells release oxygen to tissues in capillaries), the HbS molecule undergoes a conformational change into its T-state. This conformational shift is critical because it causes the newly introduced hydrophobic valine residue at β6 to become exposed on the surface of the beta-globin subunit.
- Abnormal Hydrophobic Interaction and Polymerization:
- The exposed hydrophobic Valine at β6 on one deoxy-HbS molecule fits precisely into a complementary hydrophobic pocket on an adjacent deoxy-HbS molecule, specifically within the α-chain of another hemoglobin tetramer.
- This sets off a cascade of abnormal hydrophobic interactions between multiple deoxy-HbS molecules. These weak, non-covalent interactions drive the spontaneous self-assembly and polymerization of deoxy-HbS into long, rigid, insoluble fibers (often referred to as "sickle hemoglobin polymers" or "tactoids"). This process represents a severe form of protein aggregation.
- Red Blood Cell Sickling:
- The accumulation of these long, stiff HbS polymers distorts the internal structure of the red blood cell.
- This causes the red blood cell to lose its characteristic biconcave disc shape and become rigid, elongated, and crescent or sickle-shaped. This is the key morphological change observed on the peripheral smear.
- Clinical Manifestations Explained by Sickling:
- Vaso-occlusive Crisis (Severe Bone Pain):
- Mechanism: The rigid, sickled red blood cells cannot readily deform to pass through narrow blood vessels, particularly the microvasculature (capillaries and venules). They tend to clump together and physically obstruct blood flow.
- Effect: This leads to ischemia (reduced blood supply) and infarction (tissue death) in the affected tissues. In this patient, the severe bone pain in his hands, feet, and sternum (common sites in children) is a direct consequence of this vaso-occlusion depriving the bone marrow and bone tissue of oxygen and nutrients. This is the hallmark "sickle cell crisis."
- Severe Anemia (Haemoglobin = 6.2 g/dL, Fatigue):
- Mechanism: Sickled red blood cells are much more fragile than normal red blood cells and have a significantly shortened lifespan (10-20 days compared to 100-120 days for normal red cells). They are prematurely destroyed by the spleen and other parts of the reticuloendothelial system (extravascular hemolysis).
- Effect: This rapid destruction (hemolysis) outpaces the bone marrow's ability to produce new red blood cells, resulting in chronic and severe anemia. The patient's fatigue is a classic symptom of reduced oxygen-carrying capacity due to anemia.
- Jaundice and Elevated Bilirubin:
- Mechanism: The accelerated breakdown of red blood cells (hemolysis) releases large amounts of hemoglobin. Hemoglobin is catabolized into heme, which is then converted into bilirubin (an orange-yellow pigment).
- Effect: The liver, even if functioning normally, can be overwhelmed by the excessive production of bilirubin, leading to its accumulation in the blood. This results in jaundice (yellowing of the skin and eyes) and elevated bilirubin levels on liver function tests.
- Vaso-occlusive Crisis (Severe Bone Pain):
(c) Discuss the role of amino acid chemistry in potential therapeutic approaches to sickle cell disease.
Role of Amino Acid Chemistry in Therapeutic Approaches to SCD:
Understanding the precise amino acid change and its chemical consequences is fundamental to designing and developing targeted therapies for SCD. Many current and emerging treatments aim to counteract the effects of the Valine substitution by modulating protein-protein interactions, altering the oxygen affinity of HbS, or promoting the production of alternative hemoglobin forms.
- Preventing HbS Polymerization (Targeting Hydrophobic Interactions):
- Principle: The core problem is the abnormal hydrophobic interaction driven by β6-Valine. Therapies can aim to interfere with this interaction.
- Approaches: Developing drugs that bind to the HbS molecule at the β6-Valine site or the complementary binding pocket.
- Example: Voxelotor (Oxbryta) is a recently approved drug that works by binding to the alpha-globin chains of HbS, stabilizing hemoglobin in its high-oxygen-affinity (R-state) conformation. By doing so, it reduces the amount of deoxy-HbS available to polymerize, thereby inhibiting sickling.
- Increasing Haemoglobin Oxygen Affinity (Modulating Allostery):
- Principle: If HbS stays oxygenated for longer, it won't deoxygenate and polymerize. The β6-Valine only becomes problematic in the deoxy-state.
- Approaches: Drugs that bind to HbS and shift its oxygen dissociation curve to the left, increasing its affinity for oxygen. Voxelotor, as mentioned above, achieves this.
- Promoting Fetal Haemoglobin (HbF) Production:
- Principle: Fetal hemoglobin (α2γ2) does not contain the β-globin chain and thus lacks the β6-Valine mutation. It does not sickle. Increasing its production dilutes HbS and prevents sickling.
- Approaches: Hydroxyurea (Hydroxycarbamide) is a small molecule that reactivates γ-globin gene expression, leading to increased HbF synthesis. The increased presence of non-sickling HbF reduces the concentration of HbS, thereby raising the critical concentration for sickling and diminishing polymerization.
- Reducing Cellular Dehydration (Modulating Ion Transport):
- Principle: Dehydration of red blood cells increases the intracellular concentration of HbS, promoting polymerization.
- Approaches: Investigational drugs that aim to inhibit ion transporters like the KCl cotransporter, thereby reducing cellular water loss.
In summary, a deep understanding of the chemical properties of amino acids and how their interactions govern protein structure and function is paramount. Therapies for SCD leverage this knowledge to develop molecules that either directly prevent the abnormal hydrophobic interactions (like Voxelotor), indirectly modify the cellular environment to reduce sickling (like Hydroxyurea), or, in the future, correct the genetic error at its source.
Clinical Case Scenario: A Progressive Cognitive Decline
Mrs. Eleanor Vance, an 82-year-old retired schoolteacher, is brought to the neurology clinic by her worried daughter. Over the past 5 years, Mrs. Vance has exhibited a gradual and progressive decline in her cognitive abilities. Initially, it was subtle memory lapses, such as forgetting names or misplacing keys. More recently, she has struggled with complex tasks like managing her finances, preparing meals, and following conversations. Her daughter reports that Mrs. Vance frequently repeats herself, gets disoriented in familiar surroundings, and occasionally exhibits mood swings and agitation. There is no history of stroke or significant head trauma. A physical and neurological examination reveals no focal deficits, but a mini-mental state examination (MMSE) score indicates significant cognitive impairment. Brain imaging (MRI) shows generalized cerebral atrophy, particularly pronounced in the hippocampus and cerebral cortex, but no evidence of tumors or vascular lesions.
Based on the clinical presentation and diagnostic findings, a presumptive diagnosis of Alzheimer's Disease is made.
Questions related to Protein Misfolding in Alzheimer's Disease:
(a) Alzheimer's Disease is characterized by the accumulation of two distinct types of protein aggregates: amyloid plaques and neurofibrillary tangles. For amyloid plaques, identify the primary protein involved, describe its origin, and explain how its misfolding and aggregation contribute to the pathology.
Primary Protein and Origin:
The primary protein involved in the formation of amyloid plaques in Alzheimer's Disease is beta-amyloid (Aβ) peptide.
Aβ is not synthesized as a standalone protein but is a small fragment (typically 38-43 amino acids long) derived from a much larger, integral transmembrane protein called the Amyloid Precursor Protein (APP). The production of Aβ occurs through the sequential proteolytic cleavage of APP by two different enzymes: β-secretase and γ-secretase. The longer form, Aβ42, is particularly prone to aggregation and is considered the more pathogenic species.
Misfolding and Aggregation and Contribution to Pathology:
Normally, Aβ peptides exist as soluble monomers. However, in AD, Aβ undergoes a critical misfolding event:
- Conformational Change: The soluble Aβ monomers transition from a predominantly alpha-helical or random coil conformation to a much more stable beta-sheet-rich structure. This change in secondary structure exposes hydrophobic residues and creates surfaces conducive to self-association.
- Aggregation Cascade: These misfolded Aβ monomers then begin to aggregate in a stepwise manner, forming small, soluble oligomers (thought to be the most neurotoxic species), which grow into protofibrils and eventually deposit extracellularly as large, insoluble amyloid fibrils, forming the macroscopic amyloid plaques.
- Contribution to Pathology: The accumulation of Aβ aggregates contributes to AD pathology by causing synaptic dysfunction, inducing neuronal toxicity and oxidative stress, triggering chronic neuroinflammation, and initiating the downstream pathology of the tau protein.
(b) For neurofibrillary tangles, identify the primary protein involved, explain the specific post-translational modification that initiates its misfolding, and describe how its aggregation leads to neuronal dysfunction.
Primary Protein Involved:
The primary protein involved in the formation of neurofibrillary tangles (NFTs) is Tau protein.
Tau is a microtubule-associated protein (MAP) that is highly abundant in neurons. Its primary physiological function is to stabilize microtubules, which are essential components of the neuronal cytoskeleton for maintaining structure and facilitating intracellular transport.
Post-Translational Modification Initiating Misfolding:
The specific post-translational modification that initiates the misfolding and subsequent aggregation of tau protein in AD is hyperphosphorylation.
In AD, tau becomes abnormally and excessively phosphorylated at multiple sites. This hyperphosphorylation causes it to detach from microtubules and undergo a conformational change, exposing regions that facilitate self-association. It then misfolds and aggregates into insoluble helical filaments, eventually forming large NFTs inside neurons.
How Aggregation Leads to Neuronal Dysfunction:
The accumulation of NFTs within neurons leads to profound neuronal dysfunction and ultimately cell death by:
- Microtubule Destabilization and Axonal Transport Impairment: The most direct consequence is the loss of tau's physiological function, leading to the breakdown of axonal transport pathways. This impairs synaptic function and leads to energy deficits and axonal degeneration.
- Sequestration of Normal Proteins: Tau aggregates can sequester normal, functional proteins, disrupting cellular processes.
- Physical Disruption: Large NFTs can physically impede cellular machinery, leading to cellular stress and apoptosis (programmed cell death).
(c) Discuss why aggregated proteins in Alzheimer's Disease are particularly problematic in post-mitotic cells like neurons, considering the cellular mechanisms for protein quality control and the consequences of their failure.
Aggregated proteins in Alzheimer's Disease are particularly problematic in post-mitotic cells like neurons due to a confluence of factors related to their unique cellular biology and the limitations of their protein quality control systems.
- Post-Mitotic Nature of Neurons (No Cell Division): Unlike many other cell types, mature neurons do not divide. This means they cannot "dilute" misfolded proteins among daughter cells. Once an aggregate forms, it persists and accumulates, becoming a chronic, lifelong burden for that irreplaceable cell.
- High Metabolic Demand and Oxidative Stress: Neurons are highly metabolically active, which inherently generates significant oxidative stress. This stress can damage proteins, making them more prone to misfolding and aggregation. - Complex Architecture and Axonal Transport Dependence: The long, complex structure of neurons depends on efficient axonal transport. Protein aggregates can physically obstruct this transport, leading to a breakdown in communication and a "dying back" of axons.
- Failure of Protein Quality Control (Proteostasis) Mechanisms: Cells have sophisticated systems (molecular chaperones, the ubiquitin-proteasome system, and autophagy) to refold or degrade misfolded proteins. In AD, the sheer volume and persistent nature of Aβ and tau aggregates progressively overwhelm these systems, creating a vicious cycle where the failure to clear aggregates leads to even greater accumulation and toxicity, ultimately causing neurodegeneration.
Biochemistry: Protein/Amino Acids Exam
Test your knowledge with these 40 questions.
Protein/Amino Acids Exam
Question 1/40
Exam Complete!
Here are your results, .
Your Score
38/40
95%
Carbohydrate Chemistry
Carbohydrates : Chemistry of Energy
CARBOHYDRATES
At their most fundamental level, carbohydrates are organic molecules composed of carbon (C), hydrogen (H), and oxygen (O) atoms. The most common and simplified general formula you'll see for carbohydrates is (CH₂O)n, where 'n' represents the number of carbon atoms, and 'n' is 3 or greater.
However, a more chemically precise definition,
Carbohydrates are polyhydroxy aldehydes or polyhydroxy ketones, or substances that yield these compounds upon hydrolysis.
Polyhydroxy: This is a critical term. "Poly-" means many, and "hydroxy" refers to the hydroxyl group (-OH). So, a polyhydroxy compound is one that contains multiple hydroxyl (-OH) groups attached to different carbon atoms. These hydroxyl groups are responsible for many of the characteristic properties of carbohydrates, such as their solubility in water and their ability to form hydrogen bonds.
Aldehyde: An aldehyde is an organic functional group characterized by a carbonyl group (C=O) where the carbon atom is bonded to at least one hydrogen atom and one other carbon atom (or a second hydrogen atom). It resides at the end of a carbon chain. Visualizing it: R-CHO where R is the rest of the carbon chain.
Ketone: A ketone is another organic functional group, also characterized by a carbonyl group (C=O), but in a ketone, the carbon atom of the carbonyl group is bonded to two other carbon atoms. It resides within a carbon chain, not at the end. Visualizing it: R-CO-R' where R and R' are the rest of the carbon chains.
Substances that yield these compounds upon hydrolysis: This part of the definition accounts for more complex carbohydrates (like disaccharides and polysaccharides). These larger molecules don't directly fit the polyhydroxy aldehyde/ketone description, but when they are broken down (hydrolyzed) by adding water, they release smaller units that do fit the description (monosaccharides).
In simpler terms: Carbohydrates are organic molecules that have several alcohol-like (-OH) groups and, in their simplest form, also contain either an aldehyde group or a ketone group.
The Origin of Carbohydrates: Photosynthesis
Photosynthesis is the process where plants use sunlight, water, and carbon dioxide to make glucose and oxygen.
Is a biological process carried out by plants, algae, and some types of bacteria.
The Reactants:
- Carbon Dioxide (CO₂): This is absorbed from the atmosphere. It provides the carbon atoms needed to build the carbohydrate structure.
- Water (H₂O): This is absorbed from the soil (by plants) or surrounding environment. It provides hydrogen and oxygen atoms.
- Sunlight: This is the energy source that drives the entire reaction. Chlorophyll (the green pigment in plants) captures this light energy.
The Equation:
The Products
- C₆H₁₂O₆: This is the chemical formula for glucose, the primary simple carbohydrate produced.
- O₂: Oxygen gas is released as a byproduct into the atmosphere.
Why is this important for us?
For plants: Glucose is their immediate energy source, and starch is how they store that energy. Cellulose forms their cell walls, giving them structure.
For animals (and humans): We are heterotrophs (meaning "other-feeders"). Because plants are autotrophs, (food makers). We cannot perform photosynthesis. We obtain our carbohydrates (and energy) by eating plants directly (e.g., fruits, vegetables, grains) or by eating animals that have eaten plants. When we consume these plant-derived carbohydrates, our digestive system breaks them down into simpler sugars (like glucose), which our cells then use for energy.
Importance of Carbohydrates
A. Biological:
Primary Energy Source for Living Organisms: Carbohydrates, particularly glucose, serve as the most immediate and readily available fuel source for nearly all living cells. Through cellular respiration, glucose is metabolized to produce ATP (adenosine triphosphate), that powers vital cellular processes such as muscle contraction, nerve impulse transmission, and active transport.
Storage Form of Energy: Allowing organisms to maintain energy reserves for periods of high demand or scarcity.
- Glycogen (Animals): In animals (including humans), excess glucose is polymerized and stored as glycogen, primarily in the liver and muscles. This acts as a rapidly mobilizable energy reserve, quickly converted back to glucose when blood sugar levels drop or during intense physical activity.
- Starch (Plants): Plants store surplus glucose as starch, a complex polysaccharide found in seeds, roots, and tubers. Starch serves as a long-term energy reserve, providing sustenance for plant growth, seed germination, and overwintering.
Structural Components: Carbohydrates provide structural integrity and protection to cells and tissues across diverse life forms.
- Cellulose (Plants): Forms the rigid cell walls of plants, providing tensile strength and structural support that allows plants to grow upright and resist external forces.
- Chitin (Insects, Fungi): This nitrogen-containing polysaccharide is a primary component of the tough exoskeletons of arthropods (insects, crustaceans) and the cell walls of fungi.
- Glycosaminoglycans (Humans/Animals): These complex polysaccharides (like hyaluronic acid, chondroitin sulfate, and heparin) are components of the extracellular matrix in connective tissues. They are highly hydrophilic and contribute to the structural integrity, elasticity, and hydration of tissues such as cartilage, skin, and blood vessels. For example, in cartilage, they provide resilience and act as shock absorbers.
Constituent of Nucleic Acids: Specific five-carbon sugars are integral to the backbone of the genetic material of all life.
- Ribose (RNA): This sugar is a key component of ribonucleic acid (RNA), which plays crucial roles in gene expression, protein synthesis, and regulation.
- Deoxyribose (DNA): A slightly modified version of ribose, deoxyribose forms the sugar-phosphate backbone of deoxyribonucleic acid (DNA), the molecule that carries the genetic instructions used in the growth, development, functioning, and reproduction of all known living organisms.
Dietary Fibre (Non-digestible Carbohydrates): Like cellulose, hemicellulose, and pectin, are not digestible by human enzymes but are essential for digestive health. Termed dietary fibre, they provide bulk to stool, aid in regular bowel movements, prevent constipation, and can contribute to gut microbiome health.
Lubrication, Cellular Intercommunication, & Immune Response: Glycoproteins and glycolipids on cell surfaces, are for cellular processes:
- Cell Recognition: They act as unique molecular "signatures" that allow cells to recognize each other, crucial for tissue formation, embryonic development, and immune surveillance.
- Cell Adhesion: They help cells bind to each other and to the extracellular matrix, stabilizing tissues.
- Lubrication: Glycosaminoglycans (like hyaluronic acid) are excellent lubricants, in joint fluid, reducing friction between bones.
- Immune Response: The carbohydrate patterns on cell surfaces help the immune system distinguish between "self" and "non-self" cells, triggering responses against pathogens or abnormal cells. Blood group antigens (A, B, O) are examples of cell-surface carbohydrates that dictate compatibility for blood transfusions.
Detoxification Role (e.g., Glucuronic Acid): Glucuronic acid, a derivative of glucose, is vital in the liver. It conjugates (attaches) to various toxic substances, drugs, and metabolic waste products, making them more water-soluble and easier for the body to excrete through urine or bile. This process is essential for clearing harmful compounds from the system.
B. Industrial and Commercial Applications:
- Food Industry: Used as sweeteners (e.g., sucrose, high-fructose corn syrup), thickeners (e.g., starches, gums), stabilizers, and gelling agents.
- Textile Industry: Natural fibers like cotton and linen are almost pure cellulose.
- Paper Industry: Wood pulp, rich in cellulose, is a raw material for paper production.
- Pharmaceutical Industry: Used as inactive ingredients in tablets and capsules, as drug delivery agents, and in the production of vaccines and other biologics.
- Biofuel Production: Cellulose and starch can be fermented to produce ethanol and other biofuels.
General Formula:
As mentioned, the empirical formula for many simple carbohydrates is (CH₂O)n. For example:
- Glucose, a common sugar, has the molecular formula C₆H₁₂O₆. Here, n=6, and if you divide the subscripts by 6, you get CH₂O.
- Ribose, a 5-carbon sugar, has the molecular formula C₅H₁₀O₅. Here, n=5, and again, the ratio is CH₂O.
NB: not all carbohydrates strictly adhere to this exact ratio (e.g., deoxyribose, which has one less oxygen atom than expected, or some modified carbohydrates).
Key Characteristics:
- Presence of Multiple Hydroxyl (-OH) Groups: This is the most defining feature. The abundance of these polar (-OH) groups makes carbohydrates highly hydrophilic (water-loving) and thus generally soluble in water.
- Presence of a Carbonyl (C=O) Group (Aldehyde or Ketone): This functional group (either as an aldehyde or a ketone) is what distinguishes carbohydrates chemically and dictates many of their reactions.
- Chiral Centers (Optical Centres): A chiral center (or stereocenter) is a carbon atom that is bonded to four different groups. Because most carbohydrates have multiple carbon atoms, and many of these carbons are chiral, carbohydrates can exist in various spatial arrangements called stereoisomers.
Classification of Carbohydrates
Carbohydrates are classified into groups based on the number of their constituent sugar units. The term "saccharide" (from the Greek "sakcharon" meaning sugar) is often used interchangeably with carbohydrate.
There are three primary classes of carbohydrates:
-
Monosaccharides (Simple Sugars): These are the simplest form of carbohydrates, consisting of a single polyhydroxy aldehyde or ketone unit. They are the fundamental building blocks of all carbohydrates and cannot be hydrolyzed (broken down) into simpler sugar units under mild conditions. Examples:
- Glucose: The primary metabolic fuel ("blood sugar") for most organisms.
- Fructose: Found in fruits and honey ("fruit sugar").
- Galactose: A component of lactose ("milk sugar").
- Ribose: Essential component of RNA and ATP.
- Deoxyribose: Key component of DNA.
Further Classification: Monosaccharides can be further categorized by:
- Number of Carbon Atoms: (e.g., Trioses - 3C, Pentoses - 5C like ribose, Hexoses - 6C like glucose).
- Type of Carbonyl Group: (e.g., Aldoses - containing an aldehyde group like glucose; Ketoses - containing a ketone group like fructose). For instance, glucose is an "aldohexose," and fructose is a "ketohexose."
-
Oligosaccharides: These carbohydrates are composed of a relatively small number of monosaccharide units, ranging from 2 to 10 units, linked together by glycosidic bonds. "Oligo" means "few."
Commonest Type: Disaccharides: The most prevalent type of oligosaccharide consists of two monosaccharide units joined together. Examples of Disaccharides:
- Sucrose (Table Sugar): Glucose + Fructose.
- Lactose (Milk Sugar): Glucose + Galactose.
- Maltose (Malt Sugar): Glucose + Glucose.
Formation: Disaccharides are formed by a dehydration (condensation) reaction where a water molecule is removed as two monosaccharides form a glycosidic bond.
Other Oligosaccharides (3-10 units): Examples: Raffinose (3 units - Gal-Glu-Fru), Stachyose (4 units - Gal-Gal-Glu-Fru). These are often found in legumes and can contribute to flatulence due to their non-digestibility by human enzymes until they reach gut bacteria.
-
Polysaccharides: These are large, complex carbohydrates formed by linking together many (>10, often hundreds or thousands) monosaccharide units via glycosidic bonds. "Poly" means "many."
Properties: Due to their large size, polysaccharides have a high molecular weight, are generally not sweet, and can be insoluble or form colloidal dispersions in water. Examples: Polysaccharides are diverse and can be broadly categorized by their primary biological function:
-
Storage Polysaccharides: These serve as energy reserves that can be hydrolyzed to release glucose when needed.
- Starch (Plants): The primary energy storage in plants (e.g., potatoes, grains, rice). Starch is a polymer of glucose, existing in two main forms:
- Amylose: A linear, unbranched chain of glucose
- Amylopectin: A branched polymer of glucose units,
- Glycogen (Animals): The main energy storage polysaccharide in animals, found primarily in the liver and muscles.
- Starch (Plants): The primary energy storage in plants (e.g., potatoes, grains, rice). Starch is a polymer of glucose, existing in two main forms:
- Structural Polysaccharides: These provide mechanical support, protection, and shape to cells and organisms.
-
Storage Polysaccharides: These serve as energy reserves that can be hydrolyzed to release glucose when needed.
MONOSACCHARIDES
Monosaccharides, also known as "simple sugars," are the most basic units of carbohydrates. They are single sugar molecules that cannot be hydrolyzed (broken down by water) into simpler carbohydrate units.
They serve as the primary fuel source for cells, the fundamental building blocks for more complex carbohydrates (disaccharides, oligosaccharides, and polysaccharides), and as crucial components in nucleic acids (DNA, RNA) and other vital biomolecules.
General Formula:
(CH2O)n, where 'n' usually ranges from 3 to 7, though some rarer forms can have up to 9 carbon atoms. This formula highlights that for every carbon atom, there is approximately one water molecule equivalent, hence "carbo-hydrate."
Key Characteristics & Functional Groups:
Every monosaccharide possesses defining chemical characteristics that dictate its reactivity and biological role:
One Carbonyl Group (C=O): This is the most reactive functional group and determines whether the sugar is an aldose or a ketose.
Aldehyde Group (R-CHO):If the carbonyl group is located at the end of the carbon chain (C1), it forms an aldose. Aldehyde groups are readily oxidized, making aldoses reducing sugars.Ketone Group (R-CO-R'):If the carbonyl group is located at any position other than the end of the carbon chain (typically C2 in the physiologically important ketoses), it forms a ketose. Ketones are generally less reactive than aldehydes, but ketoses can isomerize to aldoses, allowing them to also act as reducing sugars under certain conditions.
Multiple Hydroxyl Groups (-OH): At least one hydroxyl group is present on every carbon atom that doesn't bear the carbonyl group.
Polarity and Solubility:The presence of numerous highly polar hydroxyl groups makes monosaccharides exceptionally hydrophilic (water-loving) and therefore highly soluble in water. This is crucial for their transport in aqueous biological environments (e.g., blood plasma, cytoplasm).Reactivity:These hydroxyl groups are also reactive, participating in various biochemical reactions, including:- Formation of glycosidic bonds to create disaccharides and polysaccharides.
- Esterification (e.g., phosphorylation, where a phosphate group attaches to a hydroxyl group, as seen with glucose-6-phosphate).
- Oxidation (e.g., to form sugar acids) and reduction (e.g., to form sugar alcohols).
Classification of Monosaccharides:
Monosaccharides are systematically classified based on two primary structural features:
1. The Nature of the Carbonyl Group:
- Aldoses: Monosaccharides containing an aldehyde group (e.g., Glucose, Galactose, Ribose, Glyceraldehyde).
- Ketoses: Monosaccharides containing a ketone group (e.g., Fructose, Dihydroxyacetone).
2. The Number of Carbon Atoms in the Chain:
- Triose (n=3 carbons): The simplest monosaccharides.
Examples:Glyceraldehyde (an aldotriose, important in glycolysis) and Dihydroxyacetone (a ketotriose, also in glycolysis).
- Tetrose (n=4 carbons):
Examples:Erythrose (an aldotetrose, involved in pentose phosphate pathway).
- Pentose (n=5 carbons): Crucial components of nucleic acids and coenzymes.
Examples:- Ribose (an aldopentose): A key component of RNA (ribonucleic acid), ATP, and coenzymes like NAD+, FAD, and Coenzyme A.
- Deoxyribose (an aldopentose): A derivative of ribose (lacking an oxygen atom at C2), it's the sugar component of DNA (deoxyribonucleic acid).
- Xylulose & Ribulose (ketopentoses): Important intermediates in the pentose phosphate pathway.
- Hexose (n=6 carbons): The most common and physiologically significant monosaccharides, primary energy sources.
Examples:- Glucose (an aldohexose): The principal metabolic fuel for most cells, often called "blood sugar." The primary product of photosynthesis and the key starting point for cellular respiration.
- Fructose (a ketohexose): Found in fruits and honey, often called "fruit sugar." Metabolized primarily in the liver.
- Galactose (an aldohexose): Primarily found as a component of lactose (milk sugar). Converts to glucose in the liver for metabolism.
- Mannose (an aldohexose): Less common as a free sugar, but an important component of glycoproteins (proteins with attached sugars) on cell surfaces.
- Heptose (n=7 carbons):
Examples:Sedoheptulose (a ketoheptose, an intermediate in the pentose phosphate pathway).
- Octoses (n=8 carbons) & Nonoses (n=9 carbons): Rarer, but found in some bacterial cell walls and specialized biological molecules.
Combined Classification Examples:
By combining these two classification methods, we can precisely describe any monosaccharide:
- Aldotriose: A 3-carbon sugar with an aldehyde group (e.g., Glyceraldehyde).
- Ketotetrose: A 4-carbon sugar with a ketone group (e.g., Erythrulose).
- Aldopentose: A 5-carbon sugar with an aldehyde group (e.g., Ribose, Deoxyribose).
- Ketopentose: A 5-carbon sugar with a ketone group (e.g., Ribulose, Xylulose).
- Ketohexose: A 6-carbon sugar with a ketone group (e.g., Fructose).
- Aldohexose: A 6-carbon sugar with an aldehyde group (e.g., Glucose, Galactose, Mannose).
Further Detailed Aspects of Monosaccharides:
1. Stereoisomerism (Chirality)
This is a profoundly important characteristic of monosaccharides, especially for biological recognition.
- Chiral Carbons (Asymmetric Carbons): A carbon atom bonded to four different groups is called a chiral center. Monosaccharides, having multiple -OH groups, typically possess several chiral carbons.
- Enantiomers: These are stereoisomers that are non-superimposable mirror images of each other.
- D- and L- Isomers: In biochemistry, monosaccharides are primarily found in the D-configuration. This designation is based on the configuration of the chiral carbon furthest from the carbonyl group. If the -OH group on this carbon is on the right in a Fischer projection, it's a D-sugar; if it's on the left, it's an L-sugar.
- Clinical Relevance: Almost all carbohydrates used by mammalian cells are D-sugars. Enzymes are highly specific and typically only recognize and metabolize D-forms. L-forms, if present, are usually not metabolized or are excreted.
- Diastereomers: Stereoisomers that are not mirror images of each other.
- Epimers: A special type of diastereomer that differs in configuration at only one chiral carbon.
- Examples:
- Glucose and Galactose are C4 epimers (they differ only at the C4 position).
- Glucose and Mannose are C2 epimers (they differ only at the C2 position).
- Clinical Relevance: Even a single difference in the orientation of an -OH group can significantly impact how enzymes recognize and metabolize a sugar. For example, humans can metabolize glucose and galactose, but a defect in the enzyme that converts galactose to glucose can lead to galactosemia, a serious metabolic disorder.
2. Ring Formation (Cyclization)
In aqueous solutions (like within the body), monosaccharides with 5 or more carbons (and even some 4-carbon sugars) spontaneously cyclize (form rings) rather than existing as open chains. This is a crucial aspect of their structure and reactivity.
- Intramolecular Reaction: The carbonyl group (aldehyde or ketone) reacts with one of the hydroxyl groups within the same molecule.
- Hemiacetal (from aldoses): An aldehyde reacts with an alcohol.
- Hemiketal (from ketoses): A ketone reacts with an alcohol.
- Anomeric Carbon: The carbon atom that was originally the carbonyl carbon (C1 in aldoses, C2 in ketoses) becomes a new chiral center after cyclization. This carbon is called the anomeric carbon.
- Anomers (α and β): The two possible stereoisomers that can form around the anomeric carbon are called anomers.
- α-anomer: If the -OH group on the anomeric carbon is on the opposite side of the ring as the CH2OH group that defines the D/L configuration (or pointing "down" in a Haworth projection for D-sugars).
- β-anomer: If the -OH group on the anomeric carbon is on the same side of the ring as the CH2OH group (or pointing "up" in a Haworth projection for D-sugars).
- Clinical Relevance: The α or β configuration at the anomeric carbon is critical for enzyme recognition and for the type of glycosidic bonds formed in disaccharides and polysaccharides. For example, starch is made of α-glucose units, while cellulose is made of β-glucose units, and we can digest starch but not cellulose due to enzyme specificity.
- Pyranose and Furanose Rings:
- Pyranose Ring: A six-membered ring containing five carbons and one oxygen atom (e.g., α-D-glucopyranose). Glucose primarily forms pyranose rings.
- Furanose Ring: A five-membered ring containing four carbons and one oxygen atom (e.g., β-D-fructofuranose). Fructose primarily forms furanose rings, and ribose exists as a furanose in RNA.
- Equilibrium: In solution, a monosaccharide exists in an equilibrium mixture of its open-chain form and its α and β anomeric ring forms. This interconversion is called mutarotation.
Reducing Sugars
- Definition: A monosaccharide is a reducing sugar if it has a free anomeric carbon (the carbon that was part of the original aldehyde or ketone group) that can open to form an aldehyde group (even ketoses can isomerize to aldoses). This free aldehyde group can then be oxidized.
- Test: Reducing sugars can reduce oxidizing agents like Fehling's solution or Benedict's reagent (which contain Cu2+ ions) to Cu+ ions, forming a reddish precipitate.
- Clinical Relevance:
- Urinalysis for Glucose: The Benedict's test was historically used to detect glucose in urine, which is indicative of diabetes mellitus. While less common now due to more specific enzymatic tests, the principle is the same: the aldehyde group of glucose reacts.
- Glycation: In patients with uncontrolled diabetes, excess glucose in the blood can non-enzymatically react with proteins (via its free aldehyde group) in a process called glycation. This leads to the formation of Advanced Glycation End products (AGEs), which contribute to diabetic complications affecting eyes, kidneys, nerves, and blood vessels. The HbA1c test, a crucial diagnostic tool for diabetes management, measures glycated hemoglobin, reflecting average blood glucose levels over several months.
Important Monosaccharide Derivatives
Beyond the basic forms, monosaccharides can be modified for specialized roles:
- Sugar Phosphates: (e.g., Glucose-6-phosphate, Fructose-1,6-bisphosphate). Formed by adding a phosphate group to a hydroxyl group, often using ATP. These are critical intermediates in metabolic pathways (glycolysis, pentose phosphate pathway) and "trap" sugars inside the cell.
- Sugar Acids: (e.g., Gluconic acid, Glucuronic acid). Formed by the oxidation of the aldehyde or a terminal hydroxyl group. Glucuronic acid is important in detoxification pathways in the liver, conjugating with drugs and toxins to make them more water-soluble for excretion.
- Sugar Alcohols (Alditols): (e.g., Sorbitol, Xylitol). Formed by the reduction of the carbonyl group. Sorbitol can accumulate in cells of diabetic patients (e.g., in the lens of the eye), contributing to complications like cataracts. Xylitol is a common sugar substitute and has dental benefits.
- Deoxy Sugars: (e.g., 2-Deoxyribose). Lacking a hydroxyl group at one position. 2-Deoxyribose is essential for DNA.
- Amino Sugars: (e.g., Glucosamine, Galactosamine). A hydroxyl group is replaced by an amino group (NH2). These are important components of structural polysaccharides (like chitin in fungi) and glycoproteins/glycolipids (e.g., on cell surfaces, in cartilage).
Clinical Relevance
- Energy Metabolism: Glucose is the primary fuel. Understanding its structure (especially ring form and functional groups) is key to understanding how enzymes like hexokinase initiate glycolysis by phosphorylating it.
- Diabetes Mellitus: The entire disease revolves around the body's inability to regulate glucose. Knowledge of glucose's reducing properties and its ability to glycate proteins directly informs understanding of HbA1c and diabetic complications.
- Genetic Metabolic Disorders: Conditions like galactosemia (inability to metabolize galactose) or hereditary fructose intolerance (inability to metabolize fructose) arise from defects in specific enzymes that handle these monosaccharides. Early diagnosis is critical to prevent severe neurological and liver damage.
- Nutrition and Diet: Recognizing that various foods contain different monosaccharides (glucose in starchy foods, fructose in fruit, galactose in dairy) helps in dietary counseling for patients with specific metabolic needs or disorders.
- Pharmacology: Many drugs are designed to target enzymes involved in carbohydrate metabolism. For example, some anti-diabetic drugs aim to slow glucose absorption or increase its utilization.
- Cellular Recognition and Immunity: Amino sugars and other modified monosaccharides are crucial components of the glycocalyx (the carbohydrate coat on cell surfaces). These structures are vital for cell-cell recognition, adhesion, and immune responses (e.g., blood group antigens are oligosaccharides).
- Fluid and Electrolyte Balance: The high water solubility of monosaccharides means they exert osmotic pressure. In hyperglycemia, high blood glucose levels can draw water from cells into the bloodstream, leading to cellular dehydration.
Isomers: Molecules with the Same Formula, Different Structures
As established, isomers are molecules that possess the same molecular formula (meaning they have identical numbers and types of atoms) but exhibit a different arrangement of those atoms. This difference in arrangement leads to distinct chemical and/or physical properties. The existence of isomers is foundational to the vast diversity of organic molecules, particularly carbohydrates, where subtle structural differences dictate profound biological outcomes.
We categorize isomers into two primary types: Structural (Constitutional) Isomers and Stereoisomers.
1. Structural Isomers (Constitutional Isomers)
Structural isomers are characterized by having the same molecular formula but a different connectivity or sequence of bonded atoms. This means the atoms are connected to each other in a fundamentally different order, resulting in different parent structures. While less common among monosaccharides themselves (due to the strict (CH2O)n formula and functional group placement rules), understanding them provides a crucial foundation.
There are three main sub-types of structural isomerism:
a. Chain Isomerism (or Skeletal Isomerism)
Definition: These isomers differ in the arrangement of the carbon skeleton itself. The carbon atoms can be arranged in a straight chain, a branched chain, or a ring.
Example (General Chemistry): For the molecular formula C4H10 (Butane):
- n-Butane (straight chain): CH3 - CH2 - CH2 - CH3
- Isobutane (2-methylpropane, branched chain):
CH3 | CH3 - CH - CH3
Both have four carbons and ten hydrogens, but their carbon backbones are arranged differently.
Relevance to Monosaccharides: Not typically observed within the monosaccharide family (e.g., you won't find a branched-chain glucose isomer that is still a 6-carbon monosaccharide), but important for understanding overall carbohydrate structure (e.g., branched vs. unbranched polysaccharides).
b. Positional Isomerism
Definition: These isomers have the same carbon skeleton and the same functional groups, but the functional group(s) or substituent(s) are located at different positions on the carbon chain.
Example 1 (General Chemistry): Butan-1-ol vs. Butan-2-ol (Molecular formula C4H10O)
- Butan-1-ol (1-butanol): The hydroxyl (-OH) group is on the first carbon. CH3 - CH2 - CH2 - CH2 - OH
- Butan-2-ol (2-butanol): The hydroxyl (-OH) group is on the second carbon.
OH | CH3 - CH - CH2 - CH3
Relevance to Monosaccharides: While the carbonyl group defines the aldose/ketose classification, the positions of hydroxyl groups define different sugars once cyclized (e.g., the position of the anomeric -OH for α/β anomers, though this is more accurately a stereoisomer difference). Phosphorylated sugars (e.g., glucose-6-phosphate vs. glucose-1-phosphate) could be considered positional isomers if viewing the phosphate as a "substituent" on the base sugar.
c. Functional Group Isomerism
Definition: These isomers have the same molecular formula but possess different functional groups. This means the atoms are connected in such a way that they form entirely different classes of compounds with distinct chemical properties.
Example 1 (General Chemistry): Ethanol vs. Dimethyl Ether (Molecular formula C2H6O)
- Ethanol (an alcohol): Contains a hydroxyl (-OH) functional group. CH3 - CH2 - OH
- Dimethyl Ether (an ether): Contains an ether (-O-) functional group. CH3 - O - CH3
These are vastly different compounds: ethanol is a liquid at room temperature, while dimethyl ether is a gas.
Example 2 (Directly Applicable to Monosaccharides): Glucose vs. Fructose (Molecular formula C6H12O6)
- Glucose (an aldose): Contains an aldehyde (-CHO) functional group.
- Fructose (a ketose): Contains a ketone (-C=O) functional group.
Biological/Clinical Significance: This is a critical distinction! While both are hexoses and primary energy sources, their initial metabolic pathways differ. Glucose enters glycolysis directly; fructose must first be converted into glycolytic intermediates, primarily in the liver. Defects in fructose metabolism (e.g., hereditary fructose intolerance) can lead to severe health issues.
2. Stereoisomers
Stereoisomers have the same molecular formula and the same connectivity (bonding sequence) of atoms, but they differ only in the 3D arrangement of their atoms in space. This spatial arrangement, or configuration, is paramount in biology because enzymes and receptors are exquisitely sensitive to the precise three-dimensional shape of molecules.
There are two major types of stereoisomerism: Geometrical Isomerism and Optical Isomerism.
a. Geometrical Isomerism (cis-trans Isomerism)
Definition: This type of isomerism arises when there is restricted rotation around a bond, most commonly a carbon-carbon double bond (C=C), or within a ring structure. The different groups attached to the carbons involved in the restricted bond can be on the same side (cis) or opposite sides (trans) of that bond.
Requirement: Each carbon in the double bond (or in the ring that restricts rotation) must be attached to two different groups.
Biological Significance: While not directly applicable to simple monosaccharides (which don't have C=C double bonds in their carbon backbone), this type of isomerism is vital in other biological molecules like:
- Unsaturated fatty acids: cis double bonds introduce kinks, affecting membrane fluidity. trans fats (artificially created) have deleterious health effects.
- Vision pigments: The cis-trans isomerization of retinal is the primary event in light detection in the eye.
- Protein structure: Some amino acid residues, particularly proline, can exist in cis or trans conformations that influence protein folding.
Example (General Chemistry): 2-Butene (Molecular formula C4H8)
-
cis-2-Butene: The two methyl (CH3) groups are on the same side of the double bond.
CH3 CH3 \ / C = C / \ H H -
trans-2-Butene: The two methyl (CH3) groups are on opposite sides of the double bond.
CH3 H \ / C = C / \ H CH3
These are not interconvertible without breaking the double bond.
b. Optical Isomerism (Enantiomerism and Diastereomerism)
Optical isomerism refers to compounds that differ in their ability to rotate plane-polarized light. This property arises from the presence of chiral centers within the molecule.
- Chiral Center (or Asymmetric Carbon): A carbon atom bonded to four different groups. Monosaccharides, with their multiple hydroxyl groups, typically possess several chiral centers, leading to a rich array of optical isomers.
- Chirality: The property of a molecule (or an object) of being non-superimposable on its mirror image. This is like your left and right hand – they are mirror images but cannot be perfectly superimposed.
i. Enantiomers (or Optical Antipodes)
Definition: Stereoisomers that are non-superimposable mirror images of each other. They contain at least one chiral center.
Properties:
- Have identical physical properties (melting point, boiling point, density, solubility in non-chiral solvents) except for their interaction with plane-polarized light (they rotate it by an equal magnitude but in opposite directions).
- React identically with non-chiral reagents.
- Crucially, they react differently with other chiral molecules (e.g., enzymes, receptors). This is the basis for their differential biological activity.
D- and L- Designation: In biochemistry, the D- and L- system is universally used, especially for carbohydrates and amino acids. It relates to the configuration of the chiral carbon furthest from the primary functional group (carbonyl in sugars).
- D-Isomer: The hydroxyl group (-OH) on the chiral carbon furthest from the carbonyl is on the right in the Fischer projection. Most naturally occurring carbohydrates are D-sugars.
- L-Isomer: The hydroxyl group (-OH) on the chiral carbon furthest from the carbonyl is on the left in the Fischer projection. While rare, L-sugars exist (e.g., L-fucose in some glycoproteins).
Example (Monosaccharide): D-Glyceraldehyde vs. L-Glyceraldehyde (C3H6O3)
Glyceraldehyde has one chiral center (C2).
CHO CHO
| |
H-C-OH HO-C-H <-- Chiral carbon
| |
CH2OH CH2OH
D-Glyceraldehyde L-Glyceraldehyde
These are exact mirror images and cannot be superimposed.
Biological/Clinical Significance: Enzymes are typically specific for one enantiomeric form. For example, our digestive enzymes can break down D-glucose but not L-glucose. If we consumed L-glucose, it would pass through our digestive system largely undigested and unabsorbed, providing no caloric value. This specificity is why synthetic drugs often need to be produced as a single enantiomer to ensure efficacy and avoid side effects.
ii. Diastereomers
Definition: Stereoisomers that are not mirror images of each other. They arise in molecules with two or more chiral centers.
Properties:
- Have different physical and chemical properties (melting point, boiling point, solubilities, reactivity).
- Can be separated by conventional methods (unlike enantiomers).
Biological/Clinical Significance: The subtle differences in 3D structure between diastereomers allow for distinct recognition by biological systems. Our bodies distinguish between glucose, galactose, and mannose, even though they are all hexoses with the same functional group and formula.
Sub-types of Diastereomers (Crucial for Monosaccharides):
Epimers:
- Definition: Diastereomers that differ in configuration at only one of their multiple chiral centers.
- Biological Significance: Epimerization (the enzymatic interconversion of epimers) is a crucial metabolic process. For instance, in the liver, D-galactose is epimerized to D-glucose, allowing it to enter glycolytic pathways.
- Example: D-Glucose vs. D-Galactose
- Both have the formula C6H12O6.
- Both are aldohexoses with multiple chiral centers.
- They differ only in the configuration of the -OH group at Carbon 4.
- Therefore, D-glucose and D-galactose are C4 epimers.
CHO CHO | | H-C-OH H-C-OH | | HO-C-H HO-C-H | | H-C-OH HO-C-H <-- *Difference at C4* | | H-C-OH H-C-OH | | CH2OH CH2OH D-Glucose D-Galactose - Example 2: D-Glucose vs. D-Mannose are C2 epimers.
Anomers:
- Definition: A special type of diastereomer that occurs when a monosaccharide cyclizes (forms a ring). The new chiral center created at the former carbonyl carbon (the anomeric carbon) can have two different configurations (alpha or beta).
- Biological Significance: The α or β configuration at the anomeric carbon is absolutely critical for how polysaccharides are formed and their biological functions.
- Starch (the primary energy storage in plants, digestible by humans) consists of α-1,4 glycosidic linkages between glucose units.
- Cellulose (the main structural component of plant cell walls, indigestible by humans) consists of β-1,4 glycosidic linkages between glucose units. Our enzymes lack the ability to hydrolyze these β-linkages.
- Example: α-D-Glucose vs. β-D-Glucose
- When glucose forms a ring, the hydroxyl group on the anomeric carbon (C1) can be oriented either "down" (alpha) or "up" (beta) relative to the ring in a Haworth projection.
- These are anomers, and they are diastereomers because they are not mirror images. In solution, α-D-glucose, β-D-glucose, and a small amount of the open-chain form exist in equilibrium through mutarotation.
CH2OH CH2OH
/ /
O O
/ \ / \
C---C OH HO-C---C
| | | |
C-----C C-----C
\ / \ /
OH OH OH OH
α-D-Glucopyranose β-D-Glucopyranose
(OH on C1 is 'down') (OH on C1 is 'up')
Summary Table: Comprehensive Isomer Classification
| Isomer Type | Definition | Same Formula? | Same Connectivity? | Different 3D? | Biological Relevance/Examples |
|---|---|---|---|---|---|
| 1. Structural Isomers | |||||
| a. Chain | Different carbon skeleton arrangement | Yes | No | Yes | Less common for monosaccharides; relevant for overall polysaccharide branching. |
| b. Positional | Same skeleton & functional group, but group position differs | Yes | No | Yes | E.g., Glucose-1-phosphate vs. Glucose-6-phosphate (metabolic intermediates). |
| c. Functional | Same formula, but atoms arranged to form different functional groups | Yes | No | Yes | Glucose (aldose) vs. Fructose (ketose) – same molecular formula (C6H12O6), but different metabolic pathways, significant in diabetes and fructose intolerance. |
| 2. Stereoisomers | |||||
| a. Geometrical | Different spatial arrangement around a restricted bond (e.g., C=C, ring) | Yes | Yes | Yes | Not common in simple monosaccharides, but vital in fatty acids (cis/trans fats) and vision pigments (retinal cis/trans isomerization). |
| b. Optical | Differ in ability to rotate plane-polarized light (due to chiral centers) | Yes | Yes | Yes | Defines how molecules interact with living systems. |
| i. Enantiomers | Non-superimposable mirror images | Yes | Yes | Yes | D-sugars vs. L-sugars: Mammalian enzymes almost exclusively recognize D-sugars (e.g., D-glucose). L-sugars are often metabolically inert. Crucial for drug chirality and efficacy. |
| ii. Diastereomers | Stereoisomers that are NOT mirror images (multiple chiral centers) | Yes | Yes | Yes | Allow for distinct recognition by enzymes. |
| • Epimers | Diastereomers differing at ONLY ONE chiral center | Yes | Yes | Yes | D-Glucose vs. D-Galactose (C4 epimers), D-Glucose vs. D-Mannose (C2 epimers). Enzymatic epimerization is important for interconverting sugars in metabolism (e.g., galactose to glucose). Metabolic disorders like galactosemia stem from this. |
| • Anomers | Diastereomers formed during ring closure, differing at the anomeric carbon (C1 for aldose, C2 for ketose) | Yes | Yes | Yes | α-D-Glucose vs. β-D-Glucose. Determines the type of glycosidic bond in polysaccharides: starch (α-linkages, digestible) vs. cellulose (β-linkages, indigestible). Impacts carbohydrate digestion and fiber function. |
Common Monosaccharides: Hexoses
The most common and biologically significant monosaccharides are the hexoses, meaning they are sugars with six carbon atoms (C6H12O6). Among these, three stand out: glucose, fructose, and galactose. Their structural differences, though subtle, dictate distinct metabolic fates and clinical implications.
1. Glucose: The Body's Primary Fuel (D-Glucose)
Classification: Aldohexose (an aldehyde sugar with six carbons). Specifically, α-D-glucopyranose and β-D-glucopyranose are the most prevalent cyclic forms in solution.
Common Name: Often referred to as "grape sugar," "dextrose" (due to its dextrorotatory property, rotating plane-polarized light to the right), or most commonly, "blood sugar."
Biological Importance: The undisputed king of sugars in human metabolism.
- Primary Energy Source: Glucose is the universal and most readily available energy substrate for nearly all cells and tissues in the body.
- Brain's Obligate Fuel: The brain primarily relies on glucose for energy, consuming about 120g per day. Sustained low blood glucose (hypoglycemia) can rapidly lead to neurological dysfunction, coma, and even death.
- Red Blood Cells: Lack mitochondria, so they derive all their ATP from anaerobic glycolysis of glucose.
- Circulation and Regulation: It's the sugar circulating in our blood, maintained within a narrow concentration range (blood glucose homeostasis) by a sophisticated hormonal system involving insulin (lowers blood glucose by promoting uptake and storage) and glucagon (raises blood glucose by promoting glycogenolysis and gluconeogenesis).
- Storage: Stored in animals as glycogen (a highly branched polymer of glucose) primarily in the liver (to maintain blood glucose levels) and skeletal muscles (for local energy during contraction). In plants, it's stored as starch and forms structural cellulose.
- Building Block: A fundamental building block for many complex carbohydrates, including disaccharides (like sucrose and lactose) and polysaccharides (like starch, glycogen, and cellulose). Also a precursor for the synthesis of other sugars, amino acids, and lipids.
Clinical Significance:
- Diabetes Mellitus: The quintessential disease of glucose dysregulation, characterized by hyperglycemia (high blood glucose) due to defects in insulin production (Type 1) or action (Type 2). Understanding glucose metabolism is central to managing diabetes.
- Glycation: Elevated chronic blood glucose levels lead to non-enzymatic glycation of proteins (e.g., hemoglobin A1c, eye lens proteins, basement membrane proteins), contributing to diabetic complications like retinopathy, nephropathy, and neuropathy.
- Intravenous Dextrose: IV infusions of D5W (5% dextrose in water) or other dextrose solutions are common in clinical settings to provide energy, maintain hydration, or treat hypoglycemia.
2. Galactose: The Milk Sugar Constituent (D-Galactose)
Classification: Also an aldohexose. Specifically, a C4 epimer of D-glucose.
Common Name: Sometimes called "milk sugar" because it's a component of lactose (milk sugar).
Biological Importance:
- Component of Lactose: Galactose is rarely found free in nature in significant amounts. It's most commonly found as part of the disaccharide lactose (glucose + galactose), which is the primary carbohydrate in mammalian milk.
- Metabolism: Unlike glucose, galactose is not directly used for energy by most cells. It must be converted to glucose in the liver via a specific set of enzymes (the Leloir pathway) before it can enter the main metabolic pathways.
- Glycolipids & Glycoproteins: A critical constituent of glycolipids (e.g., cerebrosides, gangliosides in nerve cell membranes) and glycoproteins (e.g., on cell surfaces, components of the extracellular matrix). These are crucial for cell-cell recognition, communication, signal transduction, and structural integrity.
- Epimer of Glucose: As discussed, D-galactose is a C4 epimer of D-glucose. This subtle structural difference (just one -OH group orientation at C4) necessitates a distinct metabolic pathway for its conversion to glucose.
Clinical Significance:
- Galactosemia: A rare but severe inherited metabolic disorder caused by deficiencies in the enzymes of the Leloir pathway (most commonly galactose-1-phosphate uridylyltransferase). Unmetabolized galactose and its toxic byproducts (galactitol) accumulate, leading to liver damage (jaundice, hepatomegaly), cataracts, brain damage, and developmental delays. Early diagnosis and strict avoidance of lactose/galactose in the diet are life-saving.
- Lactose Intolerance: Not a problem with galactose metabolism itself, but with the inability to digest lactose due to insufficient lactase enzyme. The undigested lactose passes to the colon, causing osmotic diarrhea, gas, and bloating.
3. Fructose: The Fruit Sugar (D-Fructose)
Classification: Ketohexose (a ketone sugar with six carbons). Primarily exists in its five-membered ring form, β-D-fructofuranose, when free in solution or in disaccharides.
Common Name: Known as "fruit sugar" or "levulose" (due to its strong levorotatory property, rotating plane-polarized light to the left) because it's abundant in fruits, honey, and some vegetables.
Biological Importance:
- Sweetest Monosaccharide: Fructose is the sweetest of all naturally occurring monosaccharides, contributing significantly to the palatability of many foods and making it a common sweetener in the food industry (e.g., high-fructose corn syrup).
- Component of Sucrose: It's a key component of the disaccharide sucrose (table sugar), where it's linked to glucose via an α-1,2 glycosidic bond.
- Metabolism: Fructose is primarily metabolized in the liver (and to a lesser extent in the kidneys and small intestine). Unlike glucose, its uptake into cells (except liver) is not insulin-dependent, and its initial phosphorylation bypasses the main regulatory step of glycolysis (phosphofructokinase-1). This can lead to rapid conversion into glucose, glycogen, or fatty acids (triglycerides).
- Sperm Energy: Serves as an important energy source for sperm in seminal fluid.
Clinical Significance:
- High Fructose Intake and Metabolic Syndrome: While natural fructose in whole fruits is generally healthy, excessive consumption of added fructose (e.g., in sugary drinks, processed foods) is linked to:
- Increased Lipogenesis: Conversion of fructose into triglycerides in the liver, contributing to fatty liver disease (NAFLD) and elevated blood triglyceride levels.
- Insulin Resistance: While fructose doesn't directly stimulate insulin release, chronic high intake can contribute to overall insulin resistance.
- Uric Acid Production: Fructose metabolism generates uric acid, which can exacerbate gout and potentially contribute to hypertension.
- Hereditary Fructose Intolerance (HFI): A genetic disorder where the enzyme aldolase B, crucial for fructose metabolism in the liver, is deficient. Ingesting fructose (or sucrose) leads to a buildup of toxic intermediates, causing severe hypoglycemia, vomiting, liver failure, and kidney damage. Similar to galactosemia, strict dietary avoidance is critical.
- Fructose Malabsorption: A milder condition where intestinal cells have difficulty absorbing fructose, leading to gastrointestinal symptoms (bloating, gas, diarrhea).
Reducing Properties of Monosaccharides (Detailed)
This is a very important chemical property of monosaccharides, particularly relevant in diagnostic tests (like for diabetes) and in food chemistry.
Definition:
A reducing sugar is any sugar that is capable of acting as a reducing agent. This means it can donate electrons to another molecule, thereby reducing that molecule (and itself becoming oxidized).
Source of Reducing Power:
In the context of sugars, this reducing power comes from the presence of a free aldehyde (-CHO) group or a free ketone (C=O) group that can isomerize to an aldehyde group under certain conditions (e.g., in alkaline solutions).
Aldehyde Oxidation: The aldehyde group is easily oxidized to a carboxylic acid, releasing electrons in the process:
R-CHO (Aldehyde) + Oxidizing Agent --> R-COOH (Carboxylic Acid) + Reduced Agent
Why are Monosaccharides Reducing Sugars?
- All monosaccharides (glucose, fructose, galactose, ribose, etc.) are reducing sugars because they all possess a free aldehyde or ketone group when in their open-chain form.
- Even though monosaccharides predominantly exist in their cyclic forms in solution, there is a dynamic equilibrium with a small amount of the open-chain form. This open-chain form makes the aldehyde or ketone group available for reduction reactions.
- Ketoses as Reducing Sugars: While ketones are generally less reactive than aldehydes, ketoses like fructose can undergo tautomerization (specifically, an enediol rearrangement) in alkaline conditions to form an aldose. This allows them to also exhibit reducing properties.
Common Tests for Reducing Sugars:
These tests rely on the ability of the aldehyde group (or isomerized ketone) to reduce a metal ion, often resulting in a color change or precipitate.
Benedict's Test:
- Reagent: Benedict's solution (contains Cu2+ ions, as copper sulfate, in an alkaline medium with citrate to prevent precipitation).
- Procedure: Heat the sugar solution with Benedict's reagent.
- Result: Reducing sugars reduce the blue Cu2+ ions to brick-red Cu+ oxide (Cu2O) precipitate. The color can range from green to yellow to orange to red, depending on the concentration of the reducing sugar (more reducing sugar = more red precipitate).
- Clinical Significance: Historically, Benedict's test was used for qualitative (presence/absence) and semi-quantitative (estimation of concentration by color) detection of glucose in urine, which is a classic symptom of untreated or poorly controlled diabetes mellitus. While largely replaced by more specific enzymatic tests today, it illustrates the principle.
Fehling's Test: Similar to Benedict's test, uses Cu2+ ions in an alkaline solution (with tartrate as a chelator) to detect reducing sugars.
Biological and Clinical Significance of Reducing Properties:
Diabetes Diagnosis and Monitoring (Historical & Current):
- Urinalysis: As mentioned, the reducing property of glucose was the basis for early urine tests. Persistent glucosuria (glucose in urine) is a strong indicator of diabetes.
- Glycation (Non-Enzymatic Glycosylation): This is a critical concept in diabetes. The free aldehyde group of glucose can non-enzymatically react with the amino groups of proteins (e.g., lysine residues). This initial reversible reaction forms a Schiff base, which then rearranges to a more stable Amadori product.
- HbA1c: The most clinically significant example is the glycation of hemoglobin in red blood cells, forming glycated hemoglobin (HbA1c). The level of HbA1c reflects the average blood glucose concentration over the preceding 2-3 months. It's a gold standard for assessing long-term glycemic control in diabetic patients.
- Advanced Glycation End Products (AGEs): Further, irreversible reactions of Amadori products and other reactive carbonyls with proteins lead to the formation of AGEs. These compounds accumulate in tissues and contribute significantly to the chronic complications of diabetes, including:
- Microvascular complications: Retinopathy (eye damage), nephropathy (kidney damage), neuropathy (nerve damage).
- Macrovascular complications: Atherosclerosis (hardening of arteries), leading to heart attacks and strokes.
- Other: Cataracts, impaired wound healing.
- Fructosamine Test: Measures glycated albumin and other plasma proteins. Provides a shorter-term indicator of glycemic control (2-3 weeks) compared to HbA1c.
Food Chemistry:
- Maillard Reaction: The non-enzymatic browning of food (e.g., crust on bread, roasted coffee, seared meat) is a complex series of reactions between reducing sugars and amino acids/proteins, known as the Maillard reaction. This contributes to desirable flavors and aromas, but can also form potentially harmful compounds at high temperatures.
Renal Physiology: The renal threshold for glucose (typically 180 mg/dL or 10 mmol/L) is the plasma glucose concentration above which glucose starts to appear in the urine because the renal tubules' capacity to reabsorb glucose is saturated. Understanding the reducing nature of glucose helps explain why its presence in urine indicates a physiological abnormality.
Disaccharides & Polysaccharides
Having understood monosaccharides as individual sugar units, we now see how these units combine to form larger, molecules. This combination is facilitated by a special type of covalent bond known as the glycosidic bond.
Glycosidic Bonds:
A glycosidic bond is a covalent bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.
When two monosaccharides link together, this bond is specifically referred to as an O-glycosidic bond because it involves an oxygen atom.
Condensation Reaction
The bond forms when one monosaccharide's anomeric hydroxyl group reacts with another's hydroxyl group, releasing a water molecule (a condensation reaction).
a. Formation: Dehydration Synthesis
- Mechanism: Glycosidic bonds are formed through a dehydration synthesis (also known as a condensation reaction). In this process, a molecule of water is removed (dehydrated) when two monosaccharides combine.
- Reactants: This reaction occurs between the anomeric hydroxyl group (the -OH group on the anomeric carbon, C1 for aldoses, C2 for ketoses) of one monosaccharide and a hydroxyl group (-OH) on another carbon of a second monosaccharide.
- Reversal (Hydrolysis): The reverse reaction, hydrolysis, breaks the glycosidic bond by adding a molecule of water. This is how digestive enzymes (like amylase, lactase, sucrase) break down complex carbohydrates into their monosaccharide components.
b. Types of Glycosidic Bonds: Alpha (α) and Beta (β)
The orientation of the anomeric hydroxyl group (which dictates the configuration of the anomeric carbon) is critical in determining the type of glycosidic bond formed:
- Alpha (α) Glycosidic Bond: Formed when the anomeric hydroxyl group of the first monosaccharide is in the alpha (α) configuration (meaning it's "DOWN" in a Haworth projection) relative to the -CH2OH group (or the reference group for D-sugars) of that same sugar.
This orientation affects the overall shape and digestibility of the resulting disaccharide or polysaccharide. (Digestable) - Beta (β) Glycosidic Bond: Formed when the anomeric hydroxyl group of the first monosaccharide is in the beta (β) configuration (meaning it's "UP" in a Haworth projection) relative to the -CH2OH group (or reference group) of that same sugar. (Indigestable)
This orientation also impacts the structure and biological function.
c. Location of Glycosidic Bonds: Numbering the Carbons
To precisely describe a glycosidic bond, we specify which carbon atoms of each monosaccharide are linked. For example:
- 1→4 Glycosidic Bond: The anomeric carbon (C1) of the first monosaccharide is linked to the hydroxyl group on the C4 of the second monosaccharide.
- 1→6 Glycosidic Bond: The anomeric carbon (C1) of the first monosaccharide is linked to the hydroxyl group on the C6 of the second monosaccharide.
- 1→2 Glycosidic Bond: The anomeric carbon (C1) of the first monosaccharide is linked to the C2 of the second monosaccharide (common in sucrose, involving a ketohexose).
Disaccharides: Two Monosaccharides Joined
Disaccharides are carbohydrates formed when two monosaccharide units are joined together by a single glycosidic bond.
Three most common disaccharides:
a. Sucrose (Table Sugar)
- Composition: Glucose + Fructose
- Glycosidic Bond: α-1,2-glycosidic bond. This means the C1 of α-glucose is linked to the C2 of β-fructose. This specific linkage involves both anomeric carbons.
- Source: Abundant in sugarcane, sugar beets, fruits, and honey. It's the common "table sugar" we use daily. It's also formed naturally in plants as a primary transport sugar.
- Digestive Enzyme: Hydrolyzed by sucrase (or invertase) in the small intestine, releasing glucose and fructose for absorption.
- Reducing Property: Non-reducing sugar.
Why? The anomeric carbons of both glucose (C1) and fructose (C2) are involved in the glycosidic bond. This means neither can open up to form a free aldehyde or ketone group. Since the anomeric carbons are "locked" in the bond, sucrose cannot act as a reducing agent in typical chemical tests. - Nutritional & Clinical Significance:
- Energy: Provides a rapid source of energy upon digestion, releasing easily absorbable glucose and fructose.
- Sweetness: Contributes significantly to the palatability of foods and beverages.
- High Consumption: Excessive intake of sucrose (and other added sugars) is a major public health concern, linked to:
- Obesity: High caloric density.
- Type 2 Diabetes: Contributes to insulin resistance.
- Dental Caries: Fermented by oral bacteria, producing acids that erode tooth enamel.
- Cardiovascular Disease: High intake is correlated with increased risk factors.
b. Lactose (Milk Sugar)
- Composition: Galactose + Glucose
- Glycosidic Bond: β-1,4-glycosidic bond. This means the C1 of β-galactose is linked to the C4 of glucose. The β linkage is key here.
- Source: The primary sugar found exclusively in milk and dairy products. It's the main carbohydrate source for mammalian infants.
- Digestive Enzyme: Hydrolyzed by the enzyme lactase in the brush border of the small intestine, releasing galactose and glucose for absorption.
- Reducing Property: Reducing sugar.
Why? Although the anomeric carbon of galactose (C1) is involved in the glycosidic bond, the anomeric carbon of the glucose unit (C1) is free (not involved in the bond). This free anomeric carbon of glucose can still open up to form an aldehyde group, allowing lactose to act as a reducing agent. - Nutritional & Clinical Significance:
- Infant Nutrition: Crucial energy source for infants.
- Lactose Intolerance: A very common condition globally. Many adults experience a natural decline in lactase activity after weaning, leading to lactase non-persistence. When individuals with insufficient lactase consume lactose, it passes undigested to the colon.
- Symptoms: Colonic bacteria ferment the lactose, producing gases (hydrogen, methane, carbon dioxide) and short-chain fatty acids, leading to bloating, flatulence, abdominal pain, and osmotic diarrhea.
- Diagnosis: Hydrogen breath test (measures hydrogen gas produced by bacterial fermentation).
- Management: Dietary avoidance of lactose, use of lactase enzyme supplements, or consumption of lactose-free dairy products.
- Genetic Lactase Persistence: Some populations (e.g., of European or certain African descent) have evolved to maintain high lactase activity into adulthood, allowing them to digest lactose throughout life.
c. Maltose (Malt Sugar)
- Composition: Glucose + Glucose
- Glycosidic Bond: α-1,4-glycosidic bond. This means the C1 of one α-glucose unit is linked to the C4 of another glucose unit.
- Source: Not typically found free in large amounts in nature. It's primarily produced during the digestion of starch by enzymes like amylase (e.g., in our saliva and pancreas, during germinating seeds for brewing beer, or industrial starch hydrolysis).
- Digestive Enzyme: Hydrolyzed by maltase (also located in the brush border of the small intestine) into two glucose units.
- Reducing Property: Reducing sugar.
Why? Similar to lactose, the anomeric carbon (C1) of the second glucose unit is free (not involved in the glycosidic bond). This allows maltose to open up and exhibit reducing properties. - Nutritional & Clinical Significance:
- Starch Digestion Intermediate: A key intermediate product in the digestion of complex carbohydrates like starch and glycogen.
- Sweetener: Used in some food products and brewing due to its mild sweetness.
Summary of Reducing Properties in Disaccharides:
- Maltose and Lactose are reducing sugars because they possess a free anomeric carbon (on one of their constituent monosaccharide units) that can open up to form a free aldehyde group. This free group can then act as a reducing agent.
- Sucrose is a non-reducing sugar because the anomeric carbons of both glucose and fructose are involved in the glycosidic bond, preventing either from opening into a free aldehyde or ketone form.
Anomeric Carbon
The anomeric carbon is indeed the special carbon in a cyclic sugar that was once the aldehyde or ketone carbon in the open chain. It's unique because:
- It's the only carbon directly attached to two oxygen atoms within the ring (one from the ring oxygen and one from its own -OH group).
- Its hydroxyl group's orientation (α or β) is critical for defining the type of glycosidic bond formed when sugars link together, which in turn determines the macromolecule's structure, function, and digestibility.
- In reducing sugars, the ability of the anomeric carbon's hemiacetal/hemiketal to re-open to an aldehyde/ketone is what confers the reducing property.
Polysaccharides:
Large Polymers of MonosaccharidesPolysaccharides are long chains of monosaccharide units (ranging from hundreds to many thousands) linked together by glycosidic bonds. They are polymers, serving diverse and essential biological functions such as energy storage, structural support, cell-cell communication, and lubrication. Their complex structures arise from the type of monosaccharides, the length of the chains, the types of glycosidic bonds (α or β), and the presence of branching.
a. Homopolysaccharides: Made of a Single Type of Monosaccharide
These are polysaccharides composed of only one type of monosaccharide unit. The most common building block is glucose, but other monosaccharides can also form homopolysaccharides.
Starch (Energy Storage in Plants)
- Composition: A polymer of glucose units.
- Structure: Starch is a mixture of two types of glucose polymers, distinguished by their branching patterns:
- Amylose: An unbranched (or very sparsely branched) chain of glucose units primarily linked by α-1,4-glycosidic bonds. This linear structure tends to coil into a helix, which can trap iodine, giving a characteristic blue-black color (a common test for starch).
- Amylopectin: A branched chain of glucose units. It features a main chain linked by α-1,4-glycosidic bonds, with frequent α-1,6-glycosidic bonds at the branch points (typically every 20-30 glucose units).
- Function: The primary energy storage carbohydrate in plants (e.g., in grains like wheat, rice, corn; tubers like potatoes; and legumes). Plants store glucose in this form for later use.
- Digestibility: Easily digestible by humans and most animals due to the presence of α-glycosidic bonds. Our digestive enzymes, such as salivary amylase and pancreatic amylase, efficiently hydrolyze these bonds, breaking starch down into smaller dextrins, maltose, and ultimately glucose for absorption.
- Reducing Property: Starch (both amylose and amylopectin) is technically a reducing sugar, but its reducing power is very weak and often considered negligible in practical terms. This is because it has only one free anomeric carbon at one end of each polymer chain (the "reducing end"), while the vast majority of glucose units are involved in glycosidic bonds. Therefore, it does not typically give a positive result in common reducing sugar tests unless it has been partially broken down.
Glycogen (Energy Storage in Animals)
- Composition: A polymer of glucose units.
- Structure: Highly branched chains of glucose units, similar to amylopectin but even more extensively branched. It primarily uses α-1,4-glycosidic bonds in the main chain and highly frequent α-1,6-glycosidic bonds at branch points (typically every 8-12 glucose units). This extensive branching creates a compact structure with many non-reducing ends.
- Function: The primary energy storage carbohydrate in animals, often called "animal starch." It is predominantly stored in the liver (to maintain blood glucose homeostasis for the whole body) and skeletal muscles (to provide immediate glucose for muscle contraction).
- Digestibility: Easily digestible. When the body needs glucose, enzymes like glycogen phosphorylase rapidly cleave glucose units from the non-reducing ends, providing a quick energy supply. The high degree of branching allows for rapid mobilization of many glucose units simultaneously.
- Reducing Property: Like starch, glycogen is technically a reducing sugar (with one reducing end per molecule), but its reducing power is negligible due to its large size and limited number of free anomeric carbons relative to its mass.
- Clinical Significance:
- Glycogen Storage Diseases (GSDs): A group of genetic disorders caused by defects in the enzymes involved in glycogen synthesis or degradation. These lead to abnormal accumulation of glycogen (or abnormal glycogen structure) in various tissues, causing symptoms like hepatomegaly, hypoglycemia, and muscle weakness.
Cellulose (Structural Support in Plants)
- Composition: A polymer of glucose units.
- Structure: Unbranched, linear chains of glucose units linked by β-1,4-glycosidic bonds. This distinct β bond orientation (compared to the α bonds in starch and glycogen) causes the cellulose chains to adopt an extended, rigid conformation. Extensive hydrogen bonding between adjacent parallel chains forms strong, insoluble microfibrils, which are highly resistant to degradation.
- Function: The main structural component of plant cell walls, providing rigidity, strength, and support to plants. It is the most abundant organic polymer on Earth.
- Digestibility: Indigestible by humans. We lack the enzyme cellulase required to break the β-1,4 glycosidic bonds. Therefore, cellulose functions as dietary fiber (roughage) in the human diet, contributing to gut health, satiety, and regular bowel movements, but not providing calories. Ruminant animals (like cows, sheep) and termites can digest cellulose due to symbiotic microorganisms in their digestive tracts that produce cellulase.
- Reducing Property: Technically, cellulose has one reducing end per polymer chain, making it a very weak reducing sugar. However, due to its highly insoluble and tightly packed structure, and its enormous size, its reducing ability is practically undetectable in standard tests.
Chitin (Structural Component in Fungi and Arthropods)
- Composition: A polymer of N-acetylglucosamine units. N-acetylglucosamine is a glucose derivative where the hydroxyl group on C2 is replaced by an acetylated amino group (−NHCOCH3).
- Structure: Linear chains linked by β-1,4-glycosidic bonds, structurally similar to cellulose in its arrangement of extended parallel chains and extensive hydrogen bonding. This confers high tensile strength and rigidity.
- Function: Forms the rigid exoskeleton of insects and crustaceans (e.g., crabs, shrimp), and is a major component of the cell walls of fungi.
- Digestibility: Indigestible by humans. We lack the enzyme chitinase.
b. Heteropolysaccharides: Made of More Than One Type of Monosaccharide
These are polysaccharides composed of two or more different types of monosaccharide units. They are often more complex and include substances like glycosaminoglycans (GAGs), which are important components of connective tissues, lubricants, and the extracellular matrix (ECM).
1. Glycosaminoglycans (GAGs) - Mucopolysaccharides
What they are: Long, unbranched polysaccharide chains made of repeating disaccharide units. Each disaccharide unit typically consists of an amino sugar (like N-acetylglucosamine or N-acetylgalactosamine) and an uronic acid (like D-glucuronic acid or L-iduronic acid). They are highly negatively charged due to the presence of sulfate groups (e.g., chondroitin sulfate, keratan sulfate, heparan sulfate, dermatan sulfate) and carboxyl groups on the uronic acids.
Key Characteristics & Functions:
- Highly Negative Charge: This characteristic allows GAGs to attract and bind large amounts of water molecules.
- Viscous, Slippery Matrix: The trapped water molecules create a swollen, gel-like, and highly hydrated "ground substance" that is excellent for lubrication and shock absorption.
- Mainly Extracellular: Found predominantly outside cells, as crucial components of the connective tissues and the extracellular matrix (ECM).
Important Examples & Their Locations/Functions:
- Hyaluronic Acid (HA):
- Components: D-glucuronic acid + N-acetylglucosamine. Unique among GAGs for not containing sulfate groups and being unsulfated.
- Found in: Connective tissues (skin, synovial fluid (joints), vitreous humor (eye), umbilical cord, embryonic tissues).
- Functions: Promotes cell migration (important in wound healing, embryonic development, and also cancer metastasis), provides lubrication in joints, contributes to tissue hydration and compressibility, acts as a shock absorber. It is often injected cosmetically to reduce wrinkles.
- Heparin:
- Components: Highly sulfated repeating units, mainly D-glucuronic acid (or L-iduronic acid) and N-sulfo-D-glucosamine.
- Found in: Synthesized and stored in mast cells (immune cells), liver, lungs, skin, and found in blood.
- Function: Acts as a powerful anticoagulant (prevents blood clotting) by binding to and activating antithrombin III, which then inactivates clotting factors. Used therapeutically to prevent and treat thrombosis.
- Chondroitin Sulfate:
- Components: D-glucuronic acid + N-acetyl-D-galactosamine-4-O-sulfate (or 6-O-sulfate).
- Found in: Predominantly in cartilage, also in bone, skin, and other loose connective tissues.
- Function: Provides structural support and resilience to cartilage, contributing to its "springiness" and ability to withstand compressive forces. Often taken as a supplement for joint health, though evidence for its efficacy is mixed.
- Keratan Sulfate:
- Components: D-galactose + N-acetylglucosamine-6-O-sulfate. Unique among GAGs for containing galactose and lacking an uronic acid.
- Found in: Cornea of the eye, cartilage, bone, and loose connective tissues.
- Function: Crucial for corneal transparency and maintaining eye shape; also contributes to the hydration and structure of cartilage.
- Heparan Sulfate:
- Components: Highly varied, often L-iduronic acid (or D-glucuronic acid) + N-acetylglucosamine (or N-sulfo-D-glucosamine), with variable sulfation patterns.
- Found in: Associated with cell surfaces and basal laminae (a component of the ECM). Often linked to proteins to form heparan sulfate proteoglycans.
- Function: Acts as a co-receptor for various growth factors, cytokines, and enzymes. Involved in cell growth and differentiation, cell-cell communication, and the kidney's charge selectivity during glomerular filtration (prevents plasma proteins from leaking into urine).
- Dermatan Sulfate:
- Components: L-iduronic acid (or D-glucuronic acid) + N-acetyl-D-galactosamine-4-O-sulfate.
- Found in: Skin, blood vessels, heart valves, tendons.
- Function: Contributes to the tensile strength and elasticity of tissues.
2. Proteoglycans
What they are: These are special macromolecules where a core protein is extensively decorated with many glycosaminoglycan (GAG) chains covalently attached. They are characterized by being mostly carbohydrate by weight (often 95% carbohydrate, 5% protein).
Key Characteristics & Functions:
- Core Protein + GAGs: Imagine a central protein rod with long, bristly, highly negatively charged GAG chains extending outwards, creating a bottle-brush-like structure.
- Major Components of Extracellular Matrix (ECM): They form the hydrated "ground substance" that fills the spaces between cells and fibers (like collagen and elastin), giving tissues their structure and remarkable resilience.
- Highly Hydrated: Due to their numerous GAG chains, proteoglycans trap enormous amounts of water, creating a swollen, gel-like matrix that can withstand significant compressive forces. This "turgor" is essential for tissues like cartilage.
- Examples: Aggrecan (the major proteoglycan in cartilage, forming huge aggregates with hyaluronic acid), Decorin, Glypican, Syndecan.
- Cell-Surface Proteoglycans: Important for cell-cell communication, acting as co-receptors for growth factors, and mediating cell adhesion.
- Found in: Cartilage, connective tissues, cell surfaces, nucleus, secretory granules.
3. Glycoproteins
What they are: Proteins that have relatively short, branched carbohydrate chains (oligosaccharides) covalently attached to them. Unlike proteoglycans, the protein component is usually dominant, with carbohydrate content typically ranging from 1% to 15% by weight.
Key Characteristics & Functions:
- Protein is Dominant: The primary function is usually determined by the protein, with the carbohydrate moieties often modulating its activity, stability, or targeting.
- Diverse Functions: Involved in a vast array of biological processes.
- Cell-Cell Communication and Recognition: The carbohydrate parts act as highly specific "identification tags" on cell surfaces, crucial for cells recognizing each other (e.g., immune recognition, tissue organization).
- Receptors: Act as receptors for various ligands, including pathogens (e.g., HIV uses CXCR4 and CCR5 glycoproteins to enter cells).
- Antigens: Determine blood groups (e.g., ABO blood group antigens are specific glycoproteins/glycolipids on red blood cell surfaces).
- Immune Response: Involved in various aspects of immunity, including antibody recognition and complement activation.
- Structural Components: Found in the extracellular matrix and as components of mucus (mucin glycoproteins provide lubrication and protection to epithelial surfaces).
- Enzymes, Hormones, Transport Proteins: Many of these proteins are glycosylated, which can affect their stability, folding, secretion, and biological activity (e.g., many secreted hormones, lysosomal enzymes).
- Clinical Significance: Aberrant glycosylation patterns on glycoproteins are often observed in cancer cells, serving as diagnostic markers or targets for therapy.
Clinical Conditions Related to GAGs and Glycoproteins
- Tumors and Cancer Spread:
- Increased Hyaluronic Acid (HA) in the tumor microenvironment can facilitate cancer cell migration and metastasis.
- Changes in heparan sulfate proteoglycan expression can alter growth factor signaling, promoting uncontrolled cell proliferation and angiogenesis (new blood vessel formation to feed the tumor).
- Atherosclerosis (Hardening of Arteries):
- Abnormal accumulation and composition of GAGs (e.g., dermatan sulfate) in the arterial wall contribute to the formation of atherosclerotic plaques by affecting lipid deposition and cell adhesion.
- Arthritis and Aging:
- Osteoarthritis: Characterized by the degradation of articular cartilage. This involves the breakdown of aggrecan (a major proteoglycan) by enzymes like aggrecanase and matrix metalloproteinases (MMPs), leading to loss of cartilage integrity and joint function. Chondroitin sulfate levels and its synthesis are often affected.
- Age-related changes in GAG and proteoglycan composition can reduce the shock-absorbing capacity of connective tissues.
- Mucopolysaccharidoses (MPS - Genetic Disorders):
- A group of rare, inherited metabolic disorders caused by deficiencies in specific lysosomal enzymes responsible for the degradation of GAGs. This leads to the progressive accumulation of undegraded GAGs within lysosomes in various tissues and organs.
- Symptoms are diverse and can include skeletal deformities, coarse facial features, intellectual disability, organomegaly (enlarged liver/spleen), and cardiovascular problems (e.g., Hunter's Syndrome, Hurler's Syndrome). Early diagnosis and enzyme replacement therapy (ERT) or gene therapy can help manage symptoms.
- Pathogen Receptors:
- Many viruses and bacteria exploit cell surface glycoproteins (and sometimes glycolipids) as receptors for entry into host cells. For example, the HIV virus binds to CD4 and CCR5/CXCR4 glycoproteins on T-cells to initiate infection.
- Blood Group Antigens:
- The ABO blood group system is determined by specific oligosaccharide chains on glycoproteins and glycolipids present on the surface of red blood cells. These small carbohydrate differences are critical for blood transfusions.
- Cystic Fibrosis: Abnormalities in mucin glycoproteins (which are highly glycosylated) contribute to the thick, sticky mucus characteristic of cystic fibrosis, impairing lung and pancreatic function.
Summary Takeaways:
- Heteropolysaccharides (GAGs): Long, highly charged sugar chains (repeating disaccharide units), primarily for structure, lubrication, and shock absorption in connective tissues due to their immense water-binding capacity.
- Proteoglycans: A core protein + a multitude of large GAG chains. They form the swollen, hydrated "ground substance" of the extracellular matrix, crucial for resisting compressive forces and maintaining tissue integrity.
- Glycoproteins: A protein + relatively short, branched oligosaccharide chains. They are vital for cell recognition, signaling, immune function, and often act as receptors or provide structural roles. The carbohydrate portion fine-tunes the protein's function.
Summary Table: Disaccharides & Polysaccharides (Revised)
| Carbohydrate | Type | Monosaccharides Involved | Glycosidic Bond(s) | Reducing? (Practical) | Function/Notes |
|---|---|---|---|---|---|
| Sucrose | Disaccharide | Glucose + Fructose | α-1,2 | No | Table sugar; transport in plants; easily digestible. |
| Lactose | Disaccharide | Galactose + Glucose | β-1,4 | Yes | Milk sugar; digestion requires lactase; common intolerance. |
| Maltose | Disaccharide | Glucose + Glucose | α-1,4 | Yes | Intermediate in starch digestion; brewing. |
| Starch | Homopolysaccharide | Glucose (Amylose & Amylopectin) | α-1,4; α-1,6 (branches) | Weakly/No | Energy storage in plants; digestible by humans. |
| Glycogen | Homopolysaccharide | Glucose | α-1,4; α-1,6 (branches) | Weakly/No | Energy storage in animals (liver, muscle); highly branched for rapid glucose release. |
| Cellulose | Homopolysaccharide | Glucose | β-1,4 | No | Structural support in plants; indigestible fiber for humans. |
| Chitin | Homopolysaccharide | N-acetylglucosamine | β-1,4 | No | Structural in fungi/arthropods (exoskeletons); indigestible. |
| Hyaluronic Acid | Heteropolysaccharide | D-glucuronic acid + N-acetylglucosamine | Varied, β-linkages | No | Lubrication, shock absorption, tissue hydration; non-sulfated GAG. |
| Heparin | Heteropolysaccharide | D-glucuronic acid/L-iduronic acid + N-sulfo-D-glucosamine | Varied, α- and β-linkages | No | Anticoagulant; highly sulfated. |
| Chondroitin Sulfate | Heteropolysaccharide | D-glucuronic acid + N-acetyl-D-galactosamine-sulfate | Varied, β-linkages | No | Cartilage structure and resilience. |
| Proteoglycans | Glycoconjugate | Protein + many GAG chains | Covalent protein-GAG link | No | Major ECM component; hydration, compression resistance (e.g., Aggrecan in cartilage). |
| Glycoproteins | Glycoconjugate | Protein + few, branched oligosaccharides | Covalent protein-sugar link | No | Cell recognition, signaling, immune function, receptors (e.g., blood group antigens). |
Carbohydrates Lesson 1
Good Luck
Test your knowledge with these 51 questions.
Carbohydrates Lesson 1 Exam
Question 1/51
Exam Complete!
Here are your results, .
Your Score
48/51
94%
Vitamins Exam
Biochemistry: Vitamins Exam
Test your knowledge with these 30 questions.
Vitamins Exam
Question 1/30
Exam Complete!
Here are your results, .
Your Score
28/30
93%
Lipids Exam
Biochemistry: Lipids Exam
Test your knowledge with these 40 questions.
Lipids Exam
Question 1/40
Exam Complete!
Here are your results, .
Your Score
38/40
95%
Proteins Lesson Exam
Biochemistry: Protein/Amino Acids Exam
Test your knowledge with these 40 questions.
Protein/Amino Acids Exam
Question 1/40
Exam Complete!
Here are your results, .
Your Score
38/40
95%
Abnormal Haemoglobin: Sickle Cell Scenario
Abnormal Hemoglobin
Objectives:
- Define Abnormal Hemoglobin: Understand what constitutes "abnormal" in the context of hemoglobin structure and function.
- Classify Abnormal Hemoglobins: Categorize the main types of abnormal hemoglobins based on their molecular defects.
- Explore Structural Hemoglobinopathies:
- Examine the molecular basis of common structural variants (e.g., HbS, HbC, HbE).
- Discuss the impact of specific amino acid substitutions on hemoglobin's physical and chemical properties.
- Relate these molecular changes to the resulting clinical syndromes.
- Investigate Thalassemias (Quantitative Hemoglobinopathies):
- Differentiate between alpha (α) and beta (β) thalassemias.
- Elucidate the genetic defects leading to reduced or absent globin chain synthesis.
- Explain the pathogenic consequences of globin chain imbalance (e.g., ineffective erythropoiesis, hemolysis).
- Describe the clinical spectrum of thalassemia syndromes.
- Discuss Unstable Hemoglobins:
- Define unstable hemoglobin variants and their structural basis.
- Explain the mechanism of Heinz body formation and chronic hemolysis.
- Review Hemoglobins with Altered Oxygen Affinity:
- Explain the structural modifications that lead to increased or decreased oxygen affinity.
- Describe the clinical presentations associated with these variants (e.g., polycythemia, cyanosis).
- Summarize Diagnostic Approaches: Outline the key laboratory tests used to identify and characterize abnormal hemoglobins.
- Discuss Therapeutic Strategies: Briefly touch upon current and emerging treatments for common abnormal hemoglobin disorders.
1. Define Abnormal Hemoglobin
Abnormal hemoglobin refers to any variant of the hemoglobin molecule that deviates from the normal adult hemoglobin (HbA) in its primary amino acid sequence, structure, or quantity, leading to impaired function or stability. These abnormalities can result in a range of clinical conditions, collectively known as hemoglobinopathies, affecting the red blood cells' ability to effectively transport oxygen.
2. Classify Abnormal Hemoglobins
Abnormal hemoglobins are broadly classified based on the nature of their underlying molecular defect:
Structural Variants
(Qualitative Defects): Involve a change in the amino acid sequence of a globin chain, often from a point mutation. This results in an abnormal protein. Examples: HbS, HbC, HbE.
Thalassemias
(Quantitative Defects): Involve reduced or absent production of a structurally normal globin chain due to gene deletions or mutations. This leads to a chain imbalance. Examples: α-thalassemia, β-thalassemia.
Unstable Hemoglobins
Structural variants where an amino acid substitution destabilizes the molecule, causing it to precipitate and lead to chronic hemolysis and Heinz body formation.
Altered O₂ Affinity
Structural variants where amino acid changes affect allosteric properties, altering the ability to bind and release oxygen, leading to polycythemia or cyanosis.
3. Explore Structural Hemoglobinopathies
Structural hemoglobinopathies are characterized by the synthesis of an abnormal globin chain due to a mutation in the globin gene.
a. Hemoglobin S (HbS)
Molecular Basis: β6Glu→Val (Glutamate to Valine).
Impact: Creates a hydrophobic patch, leading to polymerization of deoxygenated HbS.
Syndrome: Sickle Cell Disease. Rigid sickled cells cause vaso-occlusion (pain crises) and chronic hemolytic anemia.
b. Hemoglobin C (HbC)
Molecular Basis: β6Glu→Lys (Glutamate to Lysine).
Impact: Reduced solubility causes HbC to crystallize within RBCs.
Syndrome: HbC Disease. Mild chronic hemolytic anemia, splenomegaly, and characteristic "target cells" on blood smear.
c. Hemoglobin E (HbE)
Molecular Basis: β26Glu→Lys (Glutamate to Lysine).
Impact: Creates an alternative mRNA splice site, causing a mild quantitative defect (thalassemic effect).
Syndrome: Mild microcytic anemia. Clinically significant when co-inherited with β-thalassemia.
4. Investigate Thalassemias (Quantitative Hemoglobinopathies)
Thalassemias are characterized by a reduced rate of synthesis or absence of one or more of the globin chains, leading to an imbalance in the production of α and β globin chains. The individual globin chains produced are structurally normal.
a. Alpha (α)-Thalassemia
Genetic Defect: Deletion of one or more of the four α-globin genes on chromosome 16.
Pathology: Excess β or γ chains form unstable tetramers (HbH, Hb Barts) that are poor oxygen carriers, leading to hemolysis and ineffective erythropoiesis.
Spectrum: Severity depends on the number of genes deleted, ranging from a silent carrier (1 gene) to fatal hydrops fetalis (4 genes).
b. Beta (β)-Thalassemia
Genetic Defect: Point mutations in the two β-globin genes on chromosome 11, reducing (β+) or eliminating (β0) synthesis.
Pathology: Excess α-chains are highly insoluble and precipitate in RBC precursors, causing severe ineffective erythropoiesis and hemolysis.
Spectrum: Ranges from asymptomatic trait (minor) to transfusion-dependent anemic (major).
5. Discuss Unstable Hemoglobins
- Definition: These are structural hemoglobin variants that have amino acid substitutions, usually in the interior hydrophobic pocket or at the heme-globin contact points, which disrupt the stability of the hemoglobin molecule.
- Structural Basis: The mutations often expose heme or critical hydrophobic regions to the aqueous environment. This leads to conformational changes that loosen the binding of heme to the globin chain.
- Mechanism of Heinz Body Formation and Chronic Hemolysis:
- The unstable hemoglobin molecules readily denature (unfold) and precipitate into insoluble aggregates.
- These precipitated, denatured hemoglobin aggregates attach to the inner surface of the red blood cell membrane, forming characteristic intracellular inclusions called Heinz bodies.
- Heinz bodies make red blood cells rigid and susceptible to removal by the spleen (extravascular hemolysis), leading to chronic hemolytic anemia.
- Examples: Hb Zurich, Hb Köln.
- Clinical Presentation: Chronic hemolytic anemia, often exacerbated by oxidative stress (e.g., certain drugs). Splenomegaly is common.
6. Review Hemoglobins with Altered Oxygen Affinity
These are structural hemoglobin variants where amino acid substitutions alter the allosteric regulation of oxygen binding and release.
a. Increased Oxygen Affinity
Mechanism: Mutations stabilize the R (oxygenated) state, making it harder to release O₂ to tissues.
Presentation (Polycythemia): Tissue hypoxia stimulates erythropoietin, leading to increased red blood cell production (erythrocytosis).
Examples: Hb Chesapeake, Hb Suresnes.
b. Decreased Oxygen Affinity
Mechanism: Mutations stabilize the T (deoxygenated) state, causing premature O₂ release.
Presentation (Cyanosis): Higher levels of deoxygenated Hb in arterial blood cause a bluish discoloration of the skin, though O₂ delivery is adequate.
Examples: Hb Kansas, Hb Beth Israel.
7. Summarize Diagnostic Approaches
The diagnosis of abnormal hemoglobin disorders relies on a combination of clinical evaluation and specialized laboratory tests:
- Complete Blood Count (CBC) with Red Blood Cell Indices: Screens for anemia, microcytosis, or polycythemia.
- Peripheral Blood Smear: Crucial for morphological assessment (sickle cells, target cells, Heinz bodies).
- Hemoglobin Electrophoresis (Alkaline & Acid pH): Separates different hemoglobin types based on their electrical charge.
- High-Performance Liquid Chromatography (HPLC): A more sensitive and quantitative method for separating hemoglobin types.
- Genetic Testing (DNA analysis): Confirms specific mutations in globin genes, essential for definitive diagnosis and prenatal screening.
- Family Studies: Screening parents and siblings can help identify carriers and clarify inheritance patterns.
- Sickling Test (Sodium Metabisulfite Test): Induces sickling of red cells containing HbS.
8. Discuss Therapeutic Strategies
Therapeutic approaches vary widely depending on the specific abnormal hemoglobin and its severity:
- Sickle Cell Disease (HbSS):
- Symptomatic Management: Pain control, hydration, transfusions.
- Disease-Modifying Therapies: Hydroxyurea (to increase HbF), L-Glutamine, Voxelotor (to prevent polymerization), Crizanlizumab (to reduce vaso-occlusion).
- Curative: Hematopoietic stem cell transplantation (HSCT), gene therapy (emerging).
- β-Thalassemia Major:
- Management: Regular blood transfusions and essential iron chelation therapy to prevent organ damage.
- Curative: HSCT, gene therapy (emerging).
- α-Thalassemia (HbH Disease): Occasional blood transfusions, folate supplementation.
- Other Variants (HbC, HbE homozygotes): Often mild and require little to no specific treatment.
- Unstable Hemoglobins: Avoidance of oxidative drugs, folate supplementation, splenectomy may be beneficial.
Analysis of Clinical Case: Sickle Cell Disease
Clinical Scenario
A 2-year-old boy from Mukono district presents with recurrent episodes of severe bone pain (hands, feet, and sternum pain), jaundice, and fatigue for 3 days.
Laboratory findings reveal:
- Haemoglobin = 6.2 g/dL (normal range: 11-16 g/dL)
- Peripheral smear: sickled red blood cells
- Liver function tests: Elevated bilirubin
- Haemoglobin electrophoresis test of his blood shows increased percentage of sickled haemoglobin (HbS)
A diagnosis of Vaso-occlusive crisis, and severe anaemia in Sickle Cell Disease was made.
- Clinical Signs: Recurrent severe bone pain (vaso-occlusive crisis), jaundice (evidence of hemolysis), and fatigue (symptom of anaemia).
- Laboratory Findings: Low haemoglobin (severe anaemia), sickled red blood cells on peripheral smear, elevated bilirubin (confirming high rate of cell breakdown), and definitive identification of sickled haemoglobin (HbS) via electrophoresis.
(a) The Amino Acid Change in Haemoglobin (HbS)
This part requires a detailed breakdown of the specific molecular error in the patient's haemoglobin protein, focusing on the identity of the amino acids and the genetic origin of the mistake.
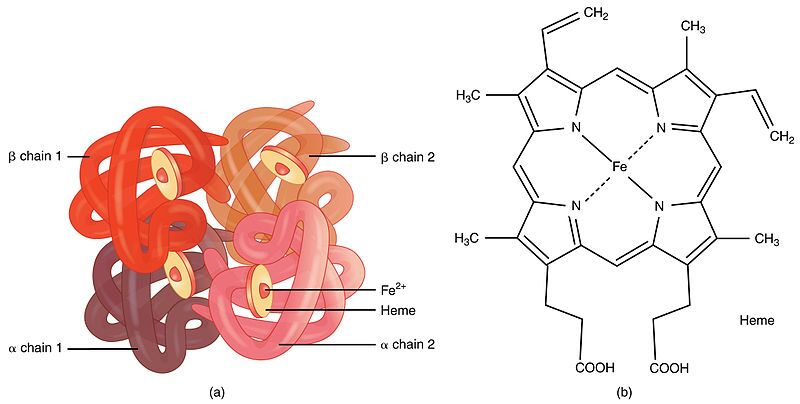
Step 1: Introduction to Haemoglobin Structure
First, it's important to understand what haemoglobin is. Haemoglobin is the primary protein found within red blood cells (erythrocytes) and its main function is to transport oxygen from the lungs to the body's tissues. It is a large, complex protein with a quaternary structure, meaning it is composed of multiple polypeptide subunits. A normal adult haemoglobin molecule (HbA) is a tetramer, consisting of four chains: two identical alpha (α)-globin chains and two identical beta (β)-globin chains. The genetic defect in sickle cell disease specifically affects the gene that provides the instructions for the beta-globin chain.

Step 2: The Specific Amino Acid Substitution
The defining molecular event in sickle cell disease is a single amino acid substitution at a precise location within the beta-globin polypeptide chain.
In a person with normal adult haemoglobin (HbA), the amino acid at the sixth position from the beginning (the N-terminus) of the beta-globin chain is Glutamic Acid (abbreviated as Glu or E).
In this patient with sickle cell disease, the haemoglobin is abnormal (called HbS). At that exact same sixth position, the Glutamic Acid has been replaced by the amino acid Valine (abbreviated as Val or V).
This single change, Glu6Val, is the sole cause of the disease.

Step 3: The Chemical Nature of the Amino Acids Involved
The severity of this substitution is due to the drastically different chemical "personalities" of the R-groups (side chains) of Glutamic Acid and Valine. This position is on the outer surface of the protein, where it is exposed to the watery environment inside the red blood cell.
| Amino Acid | Chemical Class & Properties | Behavior in Water |
|---|---|---|
| Glutamic Acid (Normal) | Its side chain contains a carboxyl group (`-CH₂-CH₂-COOH`). At the neutral pH inside a red blood cell (~7.4), this group loses a proton and becomes negatively charged (`-COO⁻`). Therefore, it is an acidic, polar, and charged amino acid. | Because it is charged and polar, Glutamic Acid is hydrophilic ("water-loving"). It forms favorable interactions with polar water molecules and is perfectly stable on the protein's surface. |
| Valine (Mutant) | Its side chain is an isopropyl group (`-CH(CH₃)₂`), which is a small, branched structure made only of carbon and hydrogen. These bonds are nonpolar. Therefore, Valine is a nonpolar, aliphatic, and neutral amino acid. | Because it is nonpolar, Valine is hydrophobic ("water-fearing"). It is thermodynamically unfavorable for this "oily" side chain to be exposed to water. It will seek to interact with other nonpolar groups to hide from the aqueous environment. |
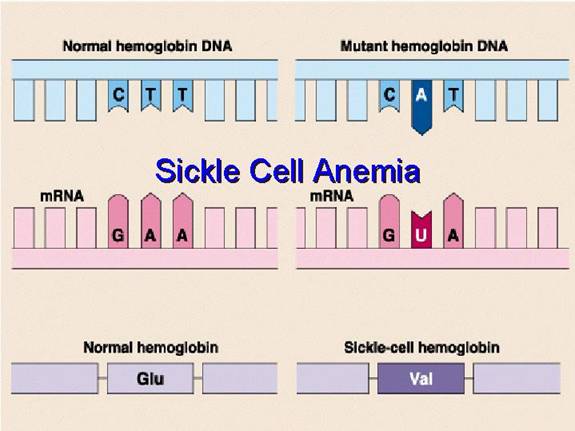
Step 4: The Chemical Basis of the Mutation (Genetics)
This amino acid error originates from a single change in the DNA sequence of the beta-globin gene. This type of mutation is called a point mutation, specifically a missense mutation because it results in a codon that codes for a different amino acid.
- The DNA Code: The genetic code is read in triplets called codons. The DNA codon on the template strand that codes for Glutamic Acid at position 6 is CTC. The corresponding codon on the coding strand is GAG.
- The Mutation: A single nucleotide change occurs where the Adenine (A) in the middle of the GAG codon is substituted for a Thymine (T). This is known as a transversion (a purine is replaced by a pyrimidine).
- Transcription to mRNA: The mutated DNA codon, now GTG on the coding strand, is transcribed into a messenger RNA (mRNA) codon. The mRNA codon becomes GUG.
- Translation to Protein: During protein synthesis at the ribosome, the cellular machinery reads the GUG codon and inserts the amino acid Valine into the growing polypeptide chain instead of Glutamic Acid.
Therefore, a single DNA base change leads to a single mRNA codon change, which in turn leads to the single, catastrophic amino acid substitution that defines sickle cell disease.
(b) Pathophysiology: From Molecular Defect to Clinical Symptoms
This section explains the step-by-step process of how the single Glu6Val substitution causes the haemoglobin to malfunction and leads to the patient's observed symptoms.

Step 1: The Molecular Effect - Polymerization of Deoxy-HbS
The key event is the behavior of HbS when it is in the deoxygenated state. In the oxygenated state (in the lungs), HbS functions almost normally as an oxygen carrier.
- Conformational Change: When a red blood cell travels to peripheral tissues and releases oxygen, the haemoglobin tetramer shifts from a high-oxygen-affinity "R-state" (relaxed) to a low-oxygen-affinity "T-state" (tense).
- Exposure of the Hydrophobic Patch: In HbS, this shift to the T-state causes a structural change that exposes the hydrophobic Valine at position β6 on the protein's surface. This creates a "sticky patch."
- Intermolecular Interaction: This exposed, oily Valine seeks to escape the aqueous cytosol. Coincidentally, the T-state conformation of another HbS molecule creates a complementary hydrophobic pocket on its surface. The Valine from one HbS molecule fits perfectly into this pocket on another HbS molecule.
- Polymerization: This initial binding is the critical step that seeds the formation of long, rigid polymers. HbS molecules begin to aggregate in a highly ordered fashion, forming long, insoluble fibers that can contain millions of haemoglobin molecules.
Step 2: The Cellular Effect - Erythrocyte Sickling
Shape Distortion: These long, stiff haemoglobin polymers grow to be longer than the diameter of the red blood cell itself. They physically push against the cell membrane from the inside, distorting the cell from its normal, flexible biconcave disc shape into a rigid, elongated, crescent or "sickle" shape.
Loss of Deformability: This sickling process causes a dramatic loss of the cell's flexibility. It becomes hard and unable to deform. This process is initially reversible if the cell becomes reoxygenated, but repeated sickling events cause permanent membrane damage, leading to irreversibly sickled cells.
Step 3: Connecting to the Clinical Manifestations
The physical properties of these sickled cells are directly responsible for the patient's symptoms:
- Vaso-occlusive Crisis (Severe Bone Pain): The rigidity and abnormal shape of the sickled cells prevent them from navigating the narrow microvasculature (capillaries). They get stuck, leading to vascular occlusion. This "logjam" blocks blood flow, causing severe tissue ischemia (lack of oxygen). The resulting hypoxia and infarction trigger intense inflammatory responses and severe pain. This is the cause of the boy's pain in his hands, feet, and sternum, which are common sites for such crises.
- Severe Anaemia (Fatigue): The sickled cells are mechanically fragile. The membrane is damaged by the internal polymers and by the stress of passing through the circulation. These cells are recognized by the reticuloendothelial system (macrophages in the spleen and liver) and are destroyed prematurely. This process, called extravascular hemolysis, reduces the average red blood cell lifespan from a normal 120 days to a mere 10-20 days. The bone marrow's production of new cells cannot keep up with this high rate of destruction, leading to a state of chronic hemolytic anaemia. The patient's very low haemoglobin level of 6.2 g/dL is a direct measure of this. The reduced oxygen-carrying capacity of the blood results in the profound fatigue.
- Jaundice (Elevated Bilirubin): The massive and continuous breakdown of red blood cells (hemolysis) leads to the release of large amounts of haemoglobin. The heme portion is catabolized into bilirubin. This high rate of bilirubin production overwhelms the liver's ability to conjugate it for excretion. The resulting buildup of unconjugated bilirubin in the bloodstream leads to hyperbilirubinemia, which manifests clinically as jaundice (yellowing of the skin and sclera), confirmed by the lab results.
(c) Therapeutic Approaches Based on Amino Acid Chemistry
Knowing that the core problem is a hydrophobic amino acid causing polymerization allows for the design of targeted therapies.
Strategy 1: Altering the Amino Acid Composition Inside the Cell
This approach aims to reduce the relative concentration of the problematic HbS.
- Induction of Fetal Haemoglobin (HbF): Fetal haemoglobin (HbF) is composed of α₂γ₂ chains. The gamma (γ)-globin chain does not have Valine at position 6 and does not participate in polymerization. Pharmacological agents like hydroxyurea can reactivate the expression of the γ-globin gene in adults. By increasing the amount of HbF inside the red blood cell, the concentration of HbS is effectively diluted. The presence of HbF molecules physically interferes with the aggregation of HbS molecules, acting as a potent polymerization inhibitor. This is a direct manipulation of the cell's overall haemoglobin amino acid profile to mitigate the effects of the faulty beta chain.
Strategy 2: Directly Targeting the Unfavorable Amino Acid Interaction
This is the most direct chemical approach, aiming to stop the Valine from interacting with its target.
- Polymerization Inhibitors: The goal is to design a molecule that prevents the key hydrophobic interaction. This can be done in several ways:
- Capping the Valine: A drug could be designed to bind directly to the exposed hydrophobic Valine at position β6, making it unavailable to interact with other molecules.
- Blocking the Pocket: A drug could bind to the complementary hydrophobic pocket on an adjacent HbS molecule, preventing the Valine from docking there.
- Altering the Conformation: A class of drugs called allosteric modulators, such as Voxelotor, binds to haemoglobin and increases its affinity for oxygen. This stabilizes the molecule in the oxygenated R-state, even at lower oxygen levels. Since polymerization only occurs in the deoxygenated T-state, this prevents the Valine from being exposed in the first place, thus inhibiting sickling. This is a therapy based entirely on manipulating the protein's shape, which is dictated by its amino acid chemistry.
Strategy 3: Correcting the Amino Acid Code at the Genetic Level
This is the most fundamental approach, aiming to fix the DNA instruction so the correct amino acid is made.
- Gene Therapy/Gene Editing: This therapeutic strategy bypasses the protein problem by going to the source. Using technologies like CRISPR-Cas9, it is possible to edit the patient's hematopoietic stem cells. The goal is to revert the mutated DNA codon GTG back to the normal GAG. By correcting the genetic blueprint, the cell's machinery will once again transcribe a GAG codon into the mRNA and translate it into Glutamic Acid. This restores the normal, hydrophilic amino acid to position 6, completely eliminating the chemical basis for polymerization and offering a potential cure for the disease.
Carbon Chemistry Lesson1 Exam
Carbon Chemistry Lesson 1
Test your knowledge with these 25 questions.
Carbon Chemistry Lesson 1
Question 1/25
Quiz Complete!
Here are your results, .
Your Score
23/25
92%
Carbohydrates First Lesson Exam
Carbohydrates Lesson 1
Good Luck
Test your knowledge with these 51 questions.
Carbohydrates Lesson 1 Exam
Question 1/51
Exam Complete!
Here are your results, .
Your Score
48/51
94%
Bioenergetics (Thermodynamics and ATP)
Bioenergetics : Thermodynamics & ATP
Bioenergetics:
The Engine of Life – How Organisms Manage Energy
Let's shift our focus now to the fundamental concept of Bioenergetics. The term itself is quite descriptive:
- "Bio" means life.
- "Energetics" means the study of energy.
Therefore, Bioenergetics is simply the study of how living organisms manage energy. It's the exploration of the transfer and utilization of energy in biological systems. It delves into the intricate mechanisms that allow life to exist and thrive, from the smallest bacteria to the largest whales.
This critical field encompasses several key aspects:
- How organisms obtain energy: This involves understanding the initial sources of energy.
- How organisms transform this energy from one form to another: Energy needs to be converted into a usable "currency".
- How organisms use this energy to do all the things necessary for life: Fueling every biological process.
At its core, Energy means the capacity or ability to do work.
In biology, "work" means anything that requires effort or causes a change. It's a much broader concept, encompassing all the dynamic processes that sustain life. Just like machines require energy to do work, all living organisms need a constant supply of energy to function.
Examples of "Work" in Biological Systems Requiring Energy:
Gross Motor Movement (Physical Work)
Just as a car needs petrol, our muscles need energy to contract and allow us to walk, run, or lift objects. Our heart muscle continuously contracts to pump blood, and our diaphragm contracts to allow us to breathe.
Growth and Development (Synthetic Work)
A toddler growing into an adult requires a massive amount of energy to synthesize new cells, tissues, and complex molecules like proteins and DNA. This is "building work." The process of reproduction also demands significant energy input.
Maintaining Homeostasis (Maintenance Work)
Even when resting, our body is still doing immense "work"! This basal metabolic activity includes our heart beating, lungs breathing, brain activity, and cells constantly repairing themselves and actively transporting ions across membranes.
Where does this Energy Come From? The Sun
For planet Earth, the main, original, and most abundant source of energy is sunlight. However, most living things, including humans, can't directly use sunlight. It's a fascinating journey through the food chain.
The Journey of Sunlight Energy to You:
-
Plants (Producers – The Solar Collectors):
Plants capture light energy from the sun through a process called photosynthesis. They use simple molecules like CO₂ and water to convert this light energy into glucose, a form of stored chemical energy. -
Animals (Consumers – The Energy Transfer Agents):
When you eat a plant product, you are directly consuming the glucose the plant made. When you eat an animal product, that animal likely consumed plants, so you are indirectly getting the energy that originally came from the sun.
The Goal: Energy-Rich Molecules for Your Cells
Regardless of what you eat, your body obtains these energy-rich molecules (carbohydrates, fats, proteins). Your cells then break them down through metabolic pathways to release the stored chemical energy. This released energy is then used to synthesize a special energy currency molecule called ATP (Adenosine Triphosphate), which is the direct fuel for almost all cellular work.
Clinical Relevance for Nursing Students
Understanding bioenergetics is foundational to many aspects of nursing:
- Nutrition and Energy Intake: Nurses assess patients' nutritional status and energy needs for processes like wound healing and recovery. Malnutrition directly impacts a patient's ability to heal.
- Metabolic Disorders: Diseases like diabetes mellitus are direct examples of impaired bioenergetics—the body's inability to properly utilize glucose for energy.
- Exercise Physiology: The energy demands of physical activity are direct applications of bioenergetics.
- Pharmacology: Many drugs affect metabolic pathways, altering how cells produce or use energy.
- Patient Education: Explaining the importance of a balanced diet can empower patients to manage their health effectively.
ATP: The Body's Universal Energy Currency
Once the body gets energy from food, it doesn't directly use these complex food molecules to power every single tiny process. Instead, the body converts the chemical energy stored in these food molecules into a much more manageable and readily available form: a special molecule called ATP.
ATP: Adenosine Triphosphate
Why is ATP called the "Energy Currency"? Think of it like money. You don't get paid in raw materials; you get paid in money, which you can use to buy whatever you need. Similarly, your body converts energy from diverse food sources into ATP (the "money"). Then, it uses ATP to "pay for" all its energy-requiring processes.
ATP is the direct, usable form of energy for almost all cellular activities.
How does ATP store and release energy?
The key lies in the "high-energy" chemical bonds connecting its three phosphate groups. When your cells need energy, they break off one of the phosphate groups from ATP. This breaking of the bond releases a significant amount of free energy that the cell can immediately use.
ATP → ADP + Pᵢ + Energy
This reaction is reversible. When your body has excess energy, it can use it to reattach the phosphate group to ADP, converting it back into ATP, thus "recharging the battery."
Free Energy (Exergonic vs. Endergonic Reactions)
Free energy (Gibbs Free Energy, G) is the amount of energy available to do work within a system. It helps us predict whether a chemical reaction will happen spontaneously (release energy) or require an input of energy.
Analogy: A person moving down a hill is a spontaneous process that releases energy. A person lifting a weight up a hill is a non-spontaneous process that requires energy.
Exergonic Reactions: Energy-Releasing
These reactions release free energy and can happen spontaneously. The change in free energy (ΔG) is negative (ΔG < 0).
Biological Examples:
- Cellular Respiration: The breakdown of glucose into CO₂ and water releases a lot of free energy used to make ATP.
- ATP Hydrolysis: The breakdown of ATP into ADP + Pᵢ is a classic exergonic reaction that releases usable energy for the cell.
Endergonic Reactions: Energy-Requiring
These reactions require an input of free energy and are non-spontaneous. The change in free energy (ΔG) is positive (ΔG > 0).
Biological Examples:
- Protein Synthesis: Building a complex protein from individual amino acids requires significant energy.
- Muscle Contraction: The process of muscle fibers shortening is an endergonic process powered by ATP.
- Active Transport: Moving substances across a cell membrane against their concentration gradient always requires energy.
The Critical Relationship: Energy Coupling
Life thrives by ingeniously linking these two types of reactions together. Cells use the energy released from an exergonic reaction (like ATP breaking down) to drive an endergonic reaction that needs energy. This is called energy coupling. ATP is the perfect intermediate, acting as the bridge that carries energy from energy-releasing pathways to energy-requiring processes.
Clinical Relevance for Nursing Students
- Cellular Function and Disease: Many diseases, like heart failure, involve inefficient energy production or utilization by cells, affecting their ability to perform endergonic work.
- Medication Impact: Some drugs target enzymes involved in ATP production or use, impacting the cell's ability to do work.
- Understanding Pathophysiology: When a patient is fatigued or weak, it often points to issues with their body's ability to generate or use ATP.
- Wound Healing and Tissue Repair: These are highly endergonic processes that require a massive input of ATP. Patients with poor nutrition or impaired energy metabolism will have difficulty healing.
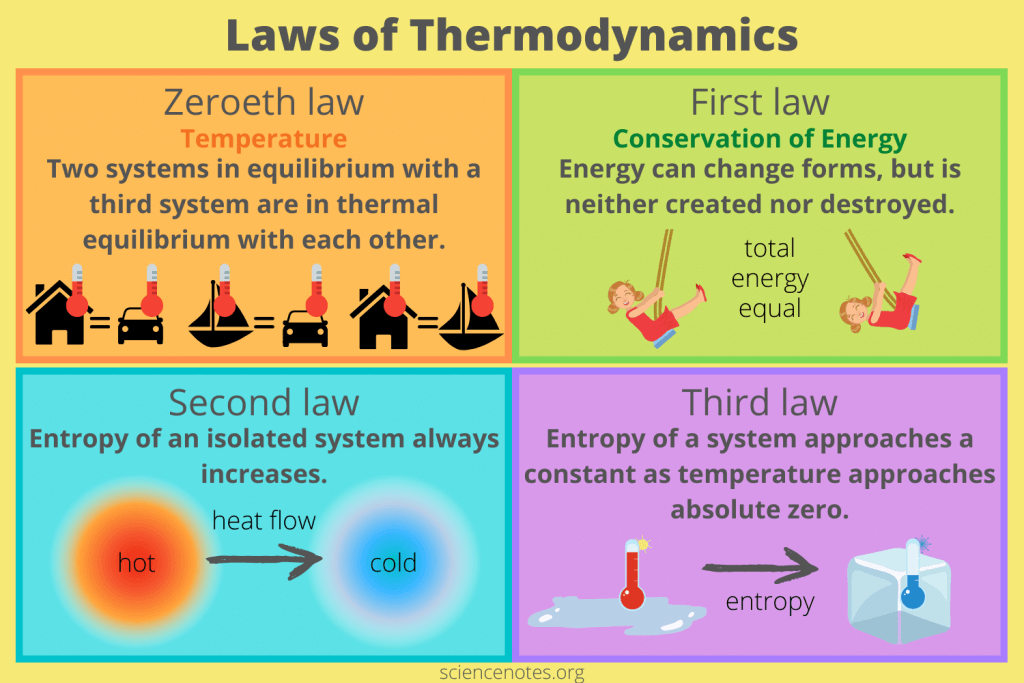
Thermodynamics:
The Universal Rules of Energy
The overarching scientific field that governs all energy concepts is Thermodynamics. It is a branch of science that deals with the transformation or interconversion of different forms of energy, and how that energy is utilized.
Literally, thermodynamics is about the power of heat or the movement of heat and energy. While "heat" is in the name, it encompasses all forms of energy relevant to biological systems, including light, thermal, chemical, electrical, and mechanical energy.
The Laws of Thermodynamics: Unbreakable Rules of the Universe
Thermodynamics is built upon a few fundamental principles known as the Laws of Thermodynamics. These laws are absolute and govern all energy transformations in the universe, including those happening inside the human body.
Zeroeth Law of Thermodynamics: Defining Temperature
"Two systems in equilibrium with a third system are in thermal equilibrium with each other."
Meaning: This law defines temperature and is the principle that allows a thermometer to accurately measure a patient's temperature.
Biological Implication: This law underpins the concept of body temperature and thermoregulation. Our bodies constantly strive to maintain a thermal equilibrium (homeostasis).
The First Law of Thermodynamics: Conservation of Energy
"Energy cannot be created or destroyed, only transformed from one form to another."
Meaning: The total amount of energy in the universe is constant. You can't get something for nothing.
Biological Implications: Plants don't "make" energy; they transform light energy into chemical energy. When you exercise, you convert chemical energy from food into mechanical energy and heat. Life needs a constant input of energy because organisms are continuously transforming it from external sources to fuel internal processes.
The Second Law of Thermodynamics: The Increase of Entropy
"In any isolated system, the total entropy (disorder) can only increase or remain constant."
Meaning: The universe naturally tends towards a state of greater disorder, randomness, or chaos. Things naturally fall apart; they do not spontaneously become more organized without external effort.
Biological Implications: Living organisms are incredibly complex, highly ordered structures. To maintain this order and fight against entropy, organisms must constantly consume energy. Life is a continuous battle against the Second Law. Every energy transformation results in some energy being "lost" as unusable heat, increasing the entropy of the environment.
Third Law of Thermodynamics: Absolute Zero and Order
"The entropy of a system approaches a constant minimum value as its temperature approaches absolute zero."
Meaning: As a system's temperature gets closer to absolute zero (-273.15 °C), the disorder of the system approaches a minimum. At absolute zero, a perfect crystal would theoretically have zero entropy (perfect order).
Biological Implication: This law highlights the relationship between temperature and molecular motion/disorder. Very low temperatures reduce molecular motion, which is why cryopreservation attempts to halt metabolic processes by drastically reducing temperature and entropy.
The Link to Bioenergetics: Thermodynamics as the Foundation
Bioenergetics is essentially the application of thermodynamic principles to biological systems. It helps us understand:
- How organisms obtain and transform energy (First Law).
- How they use energy to do work (First and Second Laws).
- Why they constantly need more energy to maintain life against the forces of entropy (Second Law).
Clinical Relevance for Nursing Students
These laws have direct clinical applications:
- Fever and Hypothermia: Understanding the Zeroeth Law contextualizes temperature regulation. A fever is an active, energy-intensive process, while hypothermia is a state where energy production cannot keep up with heat loss.
- Metabolic Rate: Basal Metabolic Rate (BMR) is a direct application of the First Law – how much energy a patient takes in versus how much they transform and use. This is crucial for managing nutrition and recovery.
- Cellular Degeneration and Aging: The Second Law helps explain the natural tendency for cells and tissues to break down over time. Aging is a continuous battle against increasing entropy.
- Wound Healing and Recovery: These processes are highly anabolic (building up), requiring significant energy input to create order (new tissue) and fight against entropy.
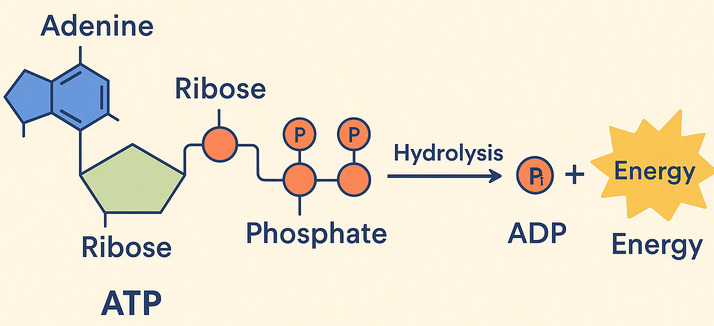
ATP: The Body's Perfect Energy Currency – A Closer Look
We've already introduced ATP as the energy currency that cells use to "pay for" their work. Now let's understand exactly how this remarkable molecule functions in this essential role.
1. The Role of "High-Energy" Bonds in ATP
ATP (Adenosine Triphosphate) is made of adenosine and three phosphate groups. The key to its power lies in the bonds between these phosphate groups, often called "high-energy phosphate bonds."
The term "high-energy" refers to the fact that when these bonds are broken, a significant amount of free energy is readily released. This is because the three negatively charged phosphate groups strongly repel each other, creating strain. Breaking the bond reduces this repulsion, and the remaining molecules (ADP and Pᵢ) settle into a more stable, lower-energy state. The difference in energy is what the cell can harness.
The Reaction: ATP Hydrolysis (Spending the Currency)
When the cell needs energy, it breaks the outermost phosphate bond in a process called hydrolysis, because a molecule of water (H₂O) is used to break the bond.
ATP + H₂O → ADP + Pᵢ + Free Energy
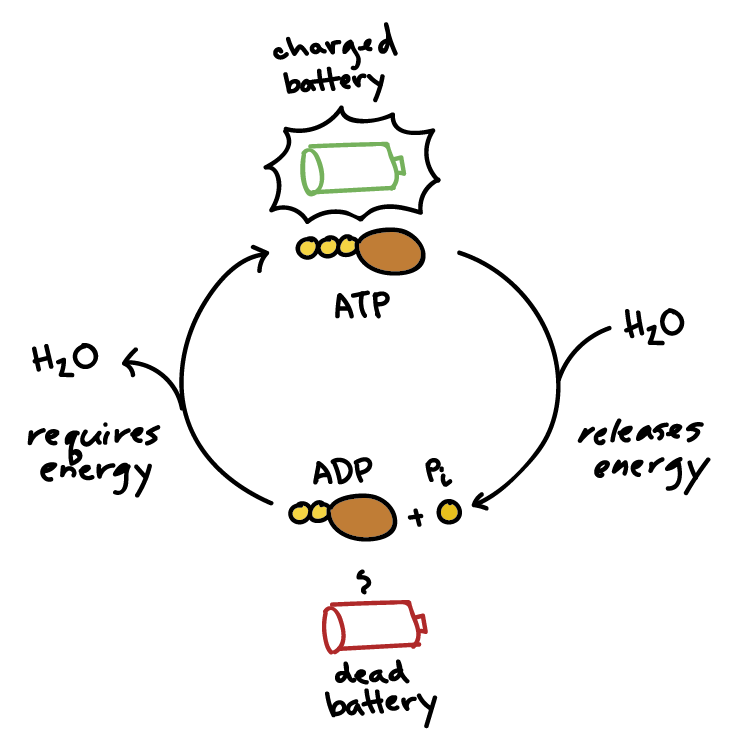
2. How Energy is "Coupled" to Power Reactions
This is truly the magic of ATP! It perfectly acts as the bridge between energy-releasing (exergonic) and energy-requiring (endergonic) processes.
The ATP Cycle & The "Coupling" Mechanism: Phosphorylation
Life depends on a continuous, rapid cycle of ATP breakdown and synthesis:
- Recharging (Endergonic): Energy from the breakdown of food is used to add a phosphate group back to ADP, reforming ATP.
ADP + Pᵢ + Energy (from food) → ATP + H₂O - Using (Exergonic): When the cell needs to perform an endergonic task, it "spends" an ATP molecule.
ATP + H₂O → ADP + Pᵢ + Free Energy (for work)
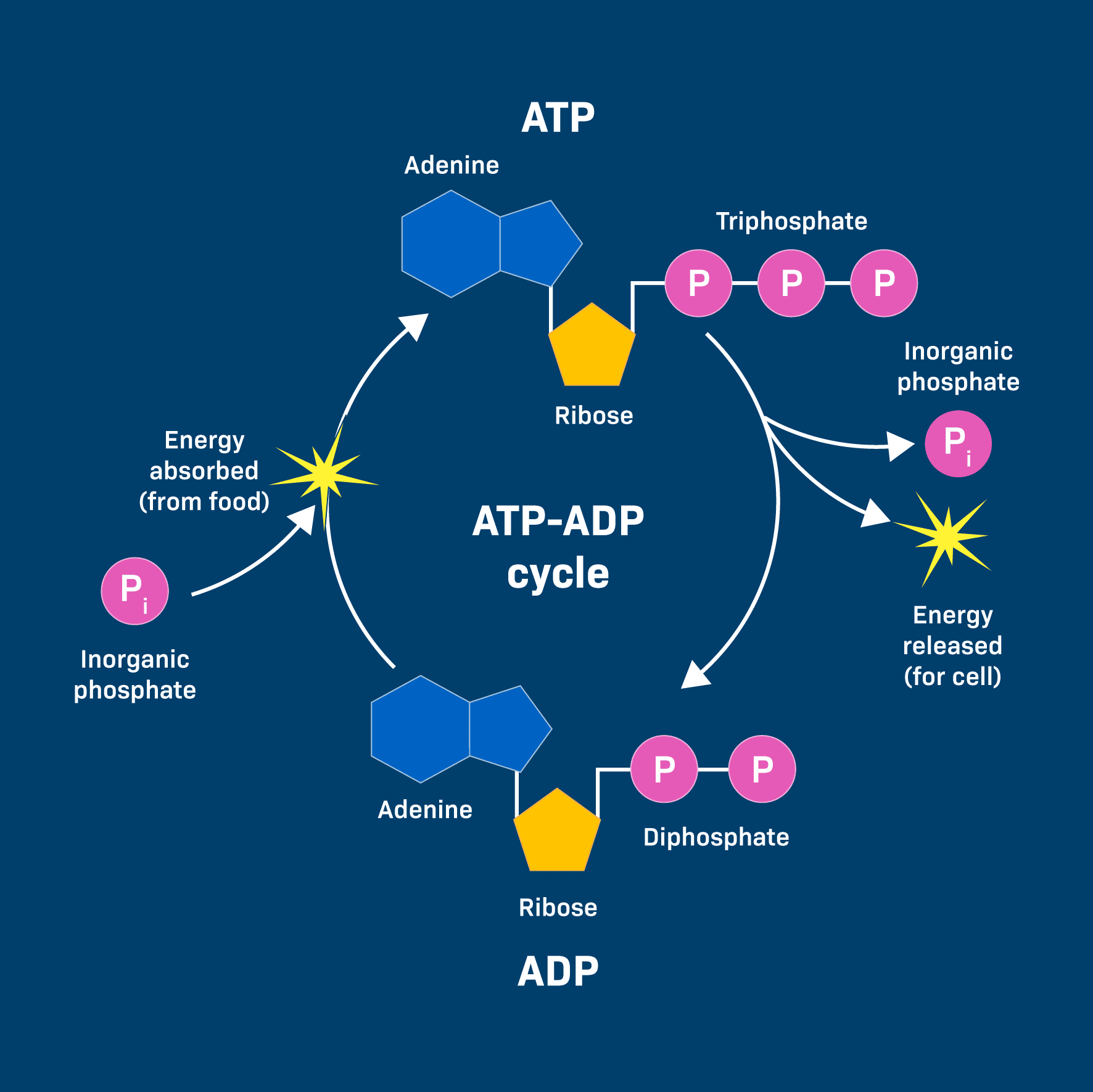
This energy is often transferred through a clever mechanism called phosphorylation. The phosphate group released from ATP is temporarily transferred to another molecule. This energizes the receiving molecule, making it more reactive and priming it to undergo its desired endergonic reaction.
Example: Muscle Contraction
An ATP molecule binds to a muscle protein (myosin). The ATP is hydrolyzed, and the phosphate (Pᵢ) temporarily attaches to the protein (phosphorylation). This causes a change in the protein's shape, leading to the physical contraction (the "work").
Why is ATP so perfect for this role?
- Manageable Energy Packet: It releases just the right amount of energy for most cellular reactions—not too much to be wasteful, and not too little to be ineffective.
- Universal Currency: Nearly all organisms and cellular processes use ATP.
- Rapid Turnover: Cells can quickly break down and resynthesize ATP, providing a constant and immediate supply of energy. A typical human adult can turn over their entire body weight in ATP every single day!
So, to summarize the continuous flow of energy that powers life:
- Sunlight → Photosynthesis (in plants) stores energy in glucose.
- Digestion breaks down glucose and other food molecules.
- Cellular Respiration (exergonic) breaks down these molecules to generate ATP from ADP.
- ATP hydrolysis (exergonic) provides bursts of free energy to power the cell's endergonic reactions (building, moving, signaling).
Clinical Relevance for Nursing Students
Understanding ATP's role is fundamental to comprehending cellular health:
- Cellular Function and Failure: When ATP production is compromised (e.g., in oxygen deprivation or metabolic poisons), cells cannot perform their essential endergonic tasks, leading to cellular damage and organ failure.
- Cardiac Function: The heart is a massive consumer of ATP. A heart attack involves a lack of oxygen, which cripples ATP production and leads to heart muscle damage.
- Pharmacology: Many drugs work by affecting ATP production or utilization pathways.
- Energy Demands of Illness: Patients recovering from surgery, trauma, or infection have significantly increased energy demands. Their bodies need to synthesize new proteins and repair tissues—all highly endergonic processes fueled by ATP.
- Diabetic Ketoacidosis (DKA): In DKA, cells cannot effectively use glucose to make ATP. They turn to fat breakdown, leading to an accumulation of ketones and acidosis, highlighting a major disruption in energy metabolism.
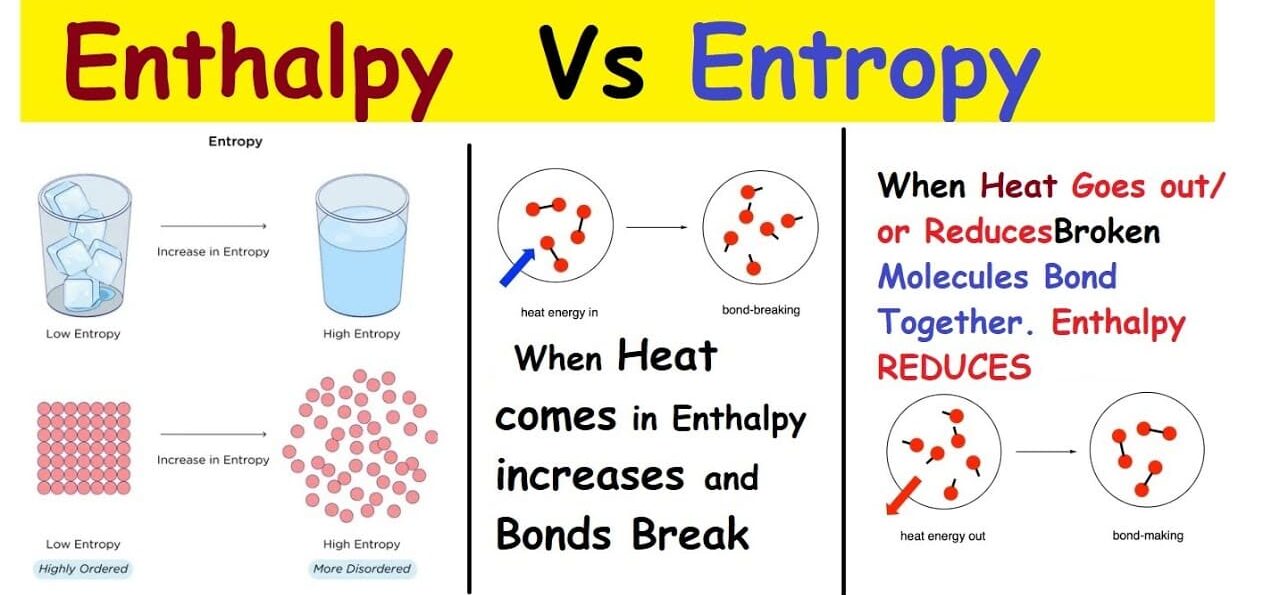
Understanding Enthalpy and Entropy (and the Gibbs Free Energy Equation)
We previously touched upon the Second Law of Thermodynamics, which introduced the powerful idea that things naturally tend towards disorder. This concept is called entropy, and it's a critical component of understanding where "free energy" comes from.
What is Entropy (ΔS)?
Entropy (S) is a fundamental thermodynamic property that serves as a quantitative measure of randomness or disorder within a system. The more ways particles can be arranged, or the more freely they can move, the higher the entropy.
Analogy: Generally, gases (high entropy, chaotic) have higher entropy than liquids (medium entropy, less ordered), which have higher entropy than solids (low entropy, ordered). Breaking large, complex molecules into smaller, simpler ones also increases entropy.
What is Enthalpy (ΔH)?
Enthalpy (H) is essentially the total heat content or the total potential energy contained within a system at constant pressure. We are most interested in the change in enthalpy (ΔH).
- Exothermic Reactions: A reaction that releases heat into the surroundings has a negative ΔH.
- Endothermic Reactions: A reaction that absorbs heat from the surroundings has a positive ΔH.
The Gibbs Free Energy Equation: ΔG = ΔH - TΔS
This powerful equation is the heart of bioenergetics because it connects these concepts to determine whether a reaction will be spontaneous (exergonic) or require energy (endergonic).
- ΔG (Change in Gibbs Free Energy): The amount of useful energy available to do work.
- A negative ΔG indicates an exergonic reaction (spontaneous, energy-releasing).
- A positive ΔG indicates an endergonic reaction (non-spontaneous, energy-requiring).
- ΔH (Change in Enthalpy): The change in total heat content. A negative ΔH (releasing heat) favors spontaneity.
- T (Temperature): The absolute temperature in Kelvin.
- ΔS (Change in Entropy): The change in disorder. A positive ΔS (increasing disorder) favors spontaneity.
Reactions are most likely to be spontaneous (exergonic) if they release heat (negative ΔH) AND increase disorder (positive ΔS).
Applying to Biological Reactions: Photosynthesis vs. Cellular Respiration
A. Photosynthesis (Endergonic)
6CO₂ + 6H₂O + Light → C₆H₁₂O₆ + 6O₂
- ΔS is negative: Simple, disordered molecules (CO₂, H₂O) are used to build a large, complex, ordered molecule (glucose). Entropy decreases.
- ΔH is positive: Energy from sunlight is absorbed to build the high-energy bonds in glucose. The reaction is endothermic.
- Overall ΔG is positive: Since both a positive ΔH and a negative ΔS make ΔG positive, photosynthesis is a highly endergonic reaction. It requires a continuous input of energy.
B. Cellular Respiration (Exergonic)
C₆H₁₂O₆ + 6O₂ → 6CO₂ + 6H₂O + Energy
- ΔS is positive: A complex, ordered molecule (glucose) is broken down into simpler, more disordered molecules (CO₂, H₂O). Entropy increases.
- ΔH is negative: The energy-rich bonds in glucose are broken, releasing stored energy as ATP and heat. The reaction is exothermic.
- Overall ΔG is negative: Since both a negative ΔH and a positive ΔS make ΔG negative, cellular respiration is a highly exergonic reaction. It releases a significant amount of free energy.
Clinical Relevance for Nursing Students
- Metabolic Rate and Heat Production: The ΔH component explains why our bodies generate heat. Cellular respiration is exothermic (negative ΔH), contributing to our core body temperature.
- Nutritional Support: Building tissues (e.g., wound healing) are endergonic processes (positive ΔG). They require significant energy input from food to proceed.
- Fever Management: Temperature (T) is a critical component of the equation. A higher T can increase the rate of some reactions but can also denature proteins if it gets too high.
- Cellular Homeostasis and Disease: Maintaining cellular order (low entropy) requires constant energy expenditure. Diseases often arise when a cell's ability to generate or use ATP breaks down, leading to an increase in cellular disorder.
- Drug Action: Some medications might influence the ΔH or ΔS of metabolic reactions, altering their favorability or rate.
Phosphoryl Group Transfers:
The Core Mechanism of ATP Energy
We've talked about ATP hydrolysis as releasing energy, but how does that energy actually get used? The primary way is through phosphoryl group transfer, often referred to simply as phosphorylation.
What is a Phosphoryl Group Transfer?
A phosphoryl group transfer is the movement of a phosphate group (Pᵢ) from one molecule to another. ATP is the most common donor. The enzyme-catalyzed transfer of the terminal phosphate group from ATP to a recipient molecule results in a phosphorylated recipient and ADP.
Why is this so effective for "energy coupling"?
- Raises the Free Energy of the Recipient: Adding a phosphate group "energizes" or "activates" the recipient molecule, making it less stable and more reactive.
- Makes Reactions More Favorable: The now-phosphorylated molecule is in a higher energy state, which can make a previously unfavorable (endergonic) reaction spontaneous.
- Induces Conformational Changes: The addition of a bulky, charged phosphate group can change a protein's shape, which is critical for processes like muscle contraction, active transport pumps, and signal transduction.
- Enzyme Regulation: Phosphorylation is a key mechanism for turning enzymes on or off.
Clinical Relevance for Nursing Students
- Cellular Function and Dysfunction: Nearly every cellular process relies on phosphoryl group transfers. Disruption of these pathways can have widespread and severe consequences.
- Drug Targets: Many drugs work by targeting enzymes involved in phosphorylation (e.g., kinase inhibitors used in cancer therapy).
- Metabolic Disorders: In conditions like diabetes, the body's ability to properly phosphorylate glucose is impaired.
Biological Oxidation-Reduction (Redox) Reactions:
The Energy Harvest
While phosphoryl group transfers are about using energy, oxidation-reduction (redox) reactions are primarily about harvesting and transferring energy from nutrient molecules. This is how cells extract energy from food.
What are Oxidation and Reduction?
These are always coupled reactions:
- Oxidation: The loss of electrons (and often hydrogen atoms).
- Reduction: The gain of electrons (and often hydrogen atoms).
A helpful mnemonic is LEO the lion says GER! (Lose Electrons Oxidation, Gain Electrons Reduction).
In biological systems, the transfer of electrons often happens along with the transfer of protons (H⁺), so oxidation often means losing hydrogen atoms (dehydrogenation), and reduction often means gaining them (hydrogenation).
Electron Carriers: The "Couriers" of Redox Energy
Cells use specialized molecules to pick up and carry electrons. The two most important are:
- NAD⁺ (Nicotinamide Adenine Dinucleotide): Its oxidized form is NAD⁺. Its reduced form, NADH, carries 2 electrons and 1 proton.
- FAD (Flavin Adenine Dinucleotide): Its oxidized form is FAD. Its reduced form, FADH₂, carries 2 electrons and 2 protons.
The Overall Flow of Energy through Redox Reactions:
- Nutrient Oxidation: Glucose is gradually oxidized in pathways like glycolysis and the Krebs cycle.
- Electron Carrier Reduction: The released electrons are picked up by NAD⁺ and FAD, reducing them to NADH and FADH₂.
- Electron Transport Chain (ETC): NADH and FADH₂ deliver these high-energy electrons to the ETC in the mitochondria.
- Energy Release: As electrons pass down a series of protein complexes, they move from a higher to a lower energy state, releasing energy.
- ATP Synthesis (Oxidative Phosphorylation): This released energy is used to pump protons, creating a gradient that drives the synthesis of large amounts of ATP.
- Oxygen as Final Electron Acceptor: At the end of the ETC, oxygen accepts the "spent" electrons and combines with protons to form water. This is why we breathe oxygen!
Clinical Relevance for Nursing Students
- Oxygen Dependence: The critical role of oxygen as the final electron acceptor highlights why hypoxia (low oxygen) severely impairs ATP synthesis, leading to cellular damage and death.
- Metabolic Poisons: Substances like cyanide block the ETC, immediately halting ATP production, which is why they are so fatal.
- Mitochondrial Diseases: Genetic disorders affecting the ETC can severely compromise a patient's ability to produce ATP, affecting high-energy organs like the brain, muscles, and heart.
- Nutritional Deficiencies: Deficiencies in vitamins that are precursors to NAD⁺ (Niacin/B3) and FAD (Riboflavin/B2) can impair electron transport and ATP production.
- Exercise and Fatigue: During intense exercise, when oxygen supply is limited, cells switch to less efficient anaerobic metabolism, which produces far less ATP.
Biochemistry Lesson Four
Bioenergetics
Test your knowledge with these 20 questions.
Biochemistry: Bioenergetics
Question 1/20
Quiz Complete!
Here are your results, .
Your Score
18/20
90%